Difference between revisions of "VMD"
m |
|||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <div id="BIO"> | ||
| + | <div class="b1"> | ||
| + | VMD | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | {Vspace} | ||
| + | |||
| + | <div class="alert"> | ||
| + | I am now using [[UCSF_Chimera|Chimera]] for all of my teaching and the VMS resources on my sites are no longer being maintained. | ||
| + | </div> | ||
| + | |||
| + | | ||
| + | | ||
| + | |||
| + | |||
[[Image:Mbp1_Model_complex.jpg|frame|none|'''A complex between a model of Mbp1 and a segment of DNA'''<br> | [[Image:Mbp1_Model_complex.jpg|frame|none|'''A complex between a model of Mbp1 and a segment of DNA'''<br> | ||
The model was visualized and positioned in VMD - nucleotides were displayed as "licorice" sticks and the protein was covered by its solvent-accessible surface, calculated with a probe radius of 1.4 Å. Coloring is ramped according to position (i.e. an atom's distance from the complex's centre of mass). The scene was rendered with the tachyon raytracer using ambient occlusion lighting and 8-fold antialiasing. ]] | The model was visualized and positioned in VMD - nucleotides were displayed as "licorice" sticks and the protein was covered by its solvent-accessible surface, calculated with a probe radius of 1.4 Å. Coloring is ramped according to position (i.e. an atom's distance from the complex's centre of mass). The scene was rendered with the tachyon raytracer using ambient occlusion lighting and 8-fold antialiasing. ]] | ||
| − | '''VMD''' ( | + | |
| + | '''VMD''' (visual molecular dynamics) is a powerful, well engineered, molecular graphics package. It is one of a number of freely available tools to visualize and manipulate three-dimensional molecular datasets, but it stands out due to its rich features, the versatility of its use and the ease with which common tasks can be accomplished and the fact that it is being actively developed and maintained. You should learn to work with it in this course, but you should also be aware that there are alternatives, each with different strengths and weaknesses. Some of the more widely used free systems are: | ||
| + | |||
| + | *[http://jmol.sourceforge.net/ JMol] | ||
| + | *[http://www.pymol.org/ PyMol] | ||
| + | *[http://spdbv.vital-it.ch/ DeepView] | ||
| + | *[http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml Cn3D] | ||
| + | *[http://www.cgl.ucsf.edu/chimera/index.html Chimera] | ||
| + | |||
| + | A comprehensive list of molecular graphics systems can be found {{WP|List_of_molecular_graphics_systems|'''here'''}}. | ||
| + | |||
| | ||
| Line 11: | Line 38: | ||
==Installing VMD== | ==Installing VMD== | ||
| + | {{task| | ||
#Access the [http://www.ks.uiuc.edu/Research/vmd/ VMD homepage] and navigate to the download section. | #Access the [http://www.ks.uiuc.edu/Research/vmd/ VMD homepage] and navigate to the download section. | ||
| − | #Follow the link to the newest version | + | #Follow the link to the newest version and review the features. |
#Access the Download page and click on the link appropriate for your platform. Follow the instructions to install VMD. | #Access the Download page and click on the link appropriate for your platform. Follow the instructions to install VMD. | ||
| + | }} | ||
| + | |||
| + | ==VMD tutorials== | ||
| + | |||
| + | The '''VMD''' site has a [http://www.ks.uiuc.edu/Research/vmd/current/docs.html#tutorials tutorial section] with many excellent contributions. | ||
| + | |||
| + | The following links take you directly to the on-line tutorials at UIUC. | ||
| + | |||
| + | *[http://www.ks.uiuc.edu/Training/Tutorials/vmd/tutorial-html/node1.html '''Part 1 (Introduction)'''] | ||
| + | |||
| + | *[http://www.ks.uiuc.edu/Training/Tutorials/vmd/tutorial-html/node2.html '''Part 2 (Working with a single molecule)'''] | ||
| + | |||
| + | |||
| + | |||
| + | <!-- | ||
| + | * [[Stereo Vision]] | ||
| + | |||
| + | |||
| + | * [[VMD_tutorial_multiple alignment|Multiple Molecules and Multiple Alignment]]== | ||
| + | ** NMR ensemble | ||
| + | ** Biotin binding site | ||
| + | |||
| + | |||
| + | * [[VMD_tutorial_analysis|Advanced Analysis of Structures]] | ||
| + | ** APBS | ||
| + | |||
| + | |||
| + | * [[VMD_tutorial_visualization|Advanced Visualization]] | ||
| + | ** Ambient occlusion lighting | ||
| + | ** Ray tracing | ||
| + | |||
| + | * [[VMD_tutorial_scripting|Scripting VMD]] | ||
| + | ** Introduction to TCL | ||
| + | ** coloring by RMSD | ||
| − | |||
| − | + | * [[VMD_tutorial_modelling|Mutation and Modeling]] | |
| + | ** ... | ||
| + | --> | ||
| − | + | ==Links and resources== | |
| − | + | *[http://www.ks.uiuc.edu/Research/vmd/ VMD homepage] | |
| − | * | + | *[http://www.ks.uiuc.edu/Research/vmd/allversions/vmd_faq.html UIUC VMD FAQ] |
| − | * | + | *[http://www.ks.uiuc.edu/Research/vmd/mailing_list/vmd-l/ VMD mailing list archive (large file!)] |
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | + | [[Category:Bioinformatics]] | |
| − | + | </div> | |
Latest revision as of 15:57, 9 June 2016
VMD
{Vspace}
I am now using Chimera for all of my teaching and the VMS resources on my sites are no longer being maintained.
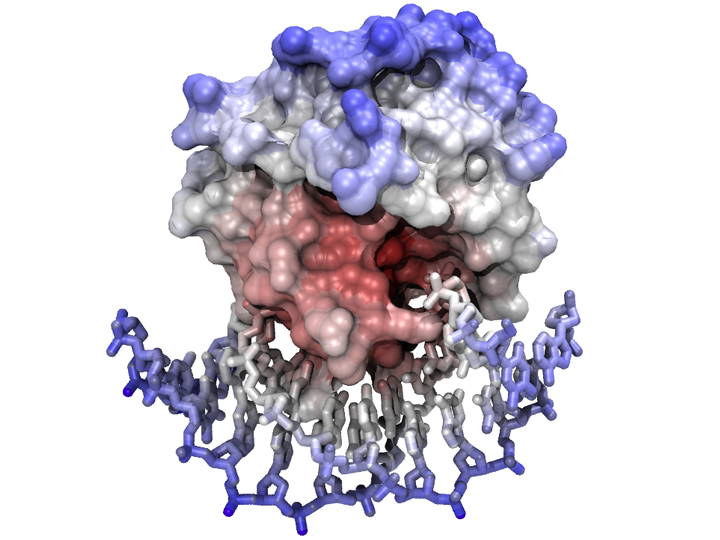
The model was visualized and positioned in VMD - nucleotides were displayed as "licorice" sticks and the protein was covered by its solvent-accessible surface, calculated with a probe radius of 1.4 Å. Coloring is ramped according to position (i.e. an atom's distance from the complex's centre of mass). The scene was rendered with the tachyon raytracer using ambient occlusion lighting and 8-fold antialiasing.
VMD (visual molecular dynamics) is a powerful, well engineered, molecular graphics package. It is one of a number of freely available tools to visualize and manipulate three-dimensional molecular datasets, but it stands out due to its rich features, the versatility of its use and the ease with which common tasks can be accomplished and the fact that it is being actively developed and maintained. You should learn to work with it in this course, but you should also be aware that there are alternatives, each with different strengths and weaknesses. Some of the more widely used free systems are:
A comprehensive list of molecular graphics systems can be found here.
Installing VMD
Task:
- Access the VMD homepage and navigate to the download section.
- Follow the link to the newest version and review the features.
- Access the Download page and click on the link appropriate for your platform. Follow the instructions to install VMD.
VMD tutorials
The VMD site has a tutorial section with many excellent contributions.
The following links take you directly to the on-line tutorials at UIUC.