Difference between revisions of "BIO Assignment Week 3"
| Line 315: | Line 315: | ||
| + | {{task|1= | ||
| − | Navigate to http://regexpal. | + | Navigate to http://regexpal.com and paste the sequence into the '''lower''' box. This site is one of a few online regular expression testers and invaluable when you are developing regular expression patterns. |
| − | Lets try | + | Lets try some expressions: |
;Most characters are matched literally. | ;Most characters are matched literally. | ||
| − | :Type " | + | :Type "<code>a</code>" in to the '''upper''' box and you will see all "<code>a</code>" characters matched. Then replace <code>a</code> with <code>q</code>. |
| − | : Now type " | + | : Now type "<code>aa</code>" instead. Then <code>krnnkk</code>. Sequences of charcters are also matched literally. |
| − | + | ;We can specify more than one character to match if we place it in square brackets. | |
| + | :<code>[lq]</code> matches <code>l</code> OR <code>q</code>. <code>[familyvw]</code> matches hydrophobic amino acids. | ||
| + | ;Within square brackets, we can specify "ranges". | ||
| + | :<code>[1-5]</code> matches digits from 1 to 5. | ||
| + | |||
| + | ;Within square brackets, we can specify characters that should NOT be matched, with the caret, <code>^</code>. | ||
| + | :<code>[^0-9]</code> matches everything EXCEPT digits. <code>[^a-z]</code> matches everything that is not a lower-case letter. | ||
| + | |||
| + | }} | ||
| + | |||
| + | One of the '''R''' functions that uses regular expressions is the function <code>gsub()</code> that replaces characters that match a "regex" with other characters. That is useful for our purpose: we can | ||
| + | #match to all characters that are NOT a letter, and | ||
| + | #replace them by - nothing: the empty string <code>""</code>. | ||
| + | This deletes them. | ||
| + | |||
| + | {{tsk|1= | ||
| + | Try this code: | ||
| + | |||
| + | <source lang="R"> | ||
| + | seq <- " | ||
| + | 1 msnqiysary sgvdvyefih stgsimkrkk ddwvnathil kaanfakakr trilekevlk | ||
| + | 61 ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf dftqtdgsas pppapkhhha | ||
| + | 121 skvdrkkair sastsaimet krnnkkaeen qfqsskilgn ptaaprkrgr pvgstrgsrr | ||
| + | 181 klgvnlqrsq sdmgfprpai pnssisttql psirstmgpq sptlgileee rhdsrqqqpq | ||
| + | 241 qnnsaqfkei dledglssdv epsqqlqqvf nqntgfvpqq qssliqtqqt esmatsvsss | ||
| + | 301 pslptspgdf adsnpfeerf pgggtspiis miprypvtsr pqtsdindkv nkylsklvdy | ||
| + | 361 fisnemksnk slpqvllhpp phsapyidap idpelhtafh wacsmgnlpi aealyeagts | ||
| + | 421 irstnsqgqt plmrsslfhn sytrrtfpri fqllhetvfd idsqsqtvih hivkrksttp | ||
| + | 481 savyyldvvl skikdfspqy rielllntqd kngdtalhia skngdvvffn tlvkmgaltt | ||
| + | 541 isnkegltan eimnqqyeqm miqngtnqhv nssntdlnih vntnnietkn dvnsmvimsp | ||
| + | 601 vspsdyityp sqiatnisrn ipnvvnsmkq masiyndlhe qhdneikslq ktlksisktk | ||
| + | 661 iqvslktlev lkesskdeng eaqtnddfei lsrlqeqntk klrkrliryk rlikqkleyr | ||
| + | 721 qtvllnklie detqattnnt vekdnntler lelaqeltml qlqrknklss lvkkfednak | ||
| + | 781 ihkyrriire gtemnieevd ssldvilqtl iannnknkga eqiitisnan sha | ||
| + | // | ||
| + | " | ||
| + | |||
| + | seq # "\n" means: line-break | ||
| + | |||
| + | seq <- gsub("[^a-zA-Z]", "", seq) | ||
| + | |||
| + | seq | ||
| + | |||
| + | # Now to change the sequence to upper-case. R has toupper() | ||
| + | # and tolower(). | ||
| + | |||
| + | toupper(seq) | ||
| + | |||
| + | # Lets put this in a function: | ||
| + | |||
| + | sanitizeSequence <- function(s) { | ||
| + | return(toupper(gsub("[^a-zA-Z]", "", s))) | ||
| + | } | ||
| + | |||
| + | # try it: | ||
| + | |||
| + | test <- "f123 a$^&&*)m. i l马马虎虎 é yßv+w" | ||
| + | sanitizeSequence(test) | ||
| + | </source> | ||
| + | |||
| + | }} | ||
| + | |||
| + | Building on this function, we can write a function to '''set''' values in our protein table. | ||
| + | |||
| + | <source lang="R"> | ||
| + | |||
| + | setDB <- function(database, | ||
| + | table, | ||
| + | id, | ||
| + | name = "", | ||
| + | refseq_id = "", | ||
| + | uniprot_id = "", | ||
| + | taxonomy_id = "", | ||
| + | genome_xref = "", | ||
| + | genome_from = "", | ||
| + | genome_to = "", | ||
| + | sequence = "", | ||
| + | species_name = "") { | ||
| + | if (missing(database) | missing(table) | missing(id)) { | ||
| + | stop("database, table and id have to be specified") | ||
| + | } | ||
| + | if (table == "protein") { | ||
| + | row <- which(database$protein$id == id) | ||
| + | if (name != "") { database$protein[row, "name"] <- name } | ||
| + | if (refseq_id != "") { database$protein[row, "refseq_id"] <- refseq_id} | ||
| + | if (uniprot_id != "") { database$protein[row, "uniprot_id"] <- uniprot_id} | ||
| + | if (taxonomy_id != "") { database$protein[row, "taxonomy_id"] <- taxonomy_id} | ||
| + | if (genome_xref != "") { database$protein[row, "genome_xref"] <- genome_xref} | ||
| + | if (genome_from != "") { database$protein[row, "genome_from"] <- genome_from} | ||
| + | if (genome_to != "") { database$protein[row, "genome_to"] <- genome_to} | ||
| + | if (sequence != "") { database$protein[row, "sequence"] <- sanitizeSequence(sequence)} | ||
| + | } | ||
| + | if (table == "taxonomy") { | ||
| + | row <- which(database$taxonomy$id == id) | ||
| + | if (species_name != "") { database$taxonomy[row, "species_name"] <- species_name } | ||
| + | } | ||
| + | return(database) | ||
| + | } | ||
| + | |||
| + | # Note that this code could be significantly improved: we are | ||
| + | # not doing any tests that the values are sane. | ||
| + | # But for now I just want to keep the code simple. | ||
| + | |||
| + | |||
| + | |||
| + | db$protein[1,] | ||
| + | |||
| + | db <- setDB(database = db, table="protein", id=1, name="Armadillo") | ||
| + | db$protein[1,] | ||
| + | db <- setDB(database = db, table="protein", id=1, name="Mbp1") | ||
| + | |||
| + | # With this, we can update our entries to add the sequence. | ||
| + | |||
| + | seq | ||
| + | |||
| + | seq <- " | ||
| + | 1 msnqiysary sgvdvyefih stgsimkrkk ddwvnathil kaanfakakr trilekevlk | ||
| + | 61 ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf dftqtdgsas pppapkhhha | ||
| + | 121 skvdrkkair sastsaimet krnnkkaeen qfqsskilgn ptaaprkrgr pvgstrgsrr | ||
| + | 181 klgvnlqrsq sdmgfprpai pnssisttql psirstmgpq sptlgileee rhdsrqqqpq | ||
| + | 241 qnnsaqfkei dledglssdv epsqqlqqvf nqntgfvpqq qssliqtqqt esmatsvsss | ||
| + | 301 pslptspgdf adsnpfeerf pgggtspiis miprypvtsr pqtsdindkv nkylsklvdy | ||
| + | 361 fisnemksnk slpqvllhpp phsapyidap idpelhtafh wacsmgnlpi aealyeagts | ||
| + | 421 irstnsqgqt plmrsslfhn sytrrtfpri fqllhetvfd idsqsqtvih hivkrksttp | ||
| + | 481 savyyldvvl skikdfspqy rielllntqd kngdtalhia skngdvvffn tlvkmgaltt | ||
| + | 541 isnkegltan eimnqqyeqm miqngtnqhv nssntdlnih vntnnietkn dvnsmvimsp | ||
| + | 601 vspsdyityp sqiatnisrn ipnvvnsmkq masiyndlhe qhdneikslq ktlksisktk | ||
| + | 661 iqvslktlev lkesskdeng eaqtnddfei lsrlqeqntk klrkrliryk rlikqkleyr | ||
| + | 721 qtvllnklie detqattnnt vekdnntler lelaqeltml qlqrknklss lvkkfednak | ||
| + | 781 ihkyrriire gtemnieevd ssldvilqtl iannnknkga eqiitisnan sha | ||
| + | // | ||
| + | " | ||
| + | |||
| + | db <- setDB(database = db, table="protein", id=1, sequence=seq) | ||
| + | |||
| + | seq <- " | ||
| + | 1 maprssavhv avysgvevye cfikgvsvmr rrrdswlnat qilkvadfdk pqrtrvlerq | ||
| + | 61 vqigahekvq ggygkyqgtw vpfqrgvdla tkykvdgims pilsldideg kaiapkkkqt | ||
| + | 121 kqkkpsvrgr rgrkpsslss stlhsvnekq pnssisptie ssmnkvnlpg aeeqvsatpl | ||
| + | 181 paspnallsp ndntikpvee lgmleapldk yeeslldffl hpeegripsf lyspppdfqv | ||
| + | 241 nsvidddght slhwacsmgh iemiklllra nadigvcnrl sqtplmrsvi ftnnydcqtf | ||
| + | 301 gqvlellqst iyavdtngqs ifhhivqsts tpskvaaaky yldcilekli siqpfenvvr | ||
| + | 361 lvnlqdsngd tslliaarng amdcvnslls ynanpsipnr qrrtaseyll eadkkphsll | ||
| + | 421 qsnsnashsa fsfsgispai ispscsshaf vkaipsissk fsqlaeeyes qlrekeedli | ||
| + | 481 ranrlkqdtl neisrtyqel tflqknnpty sqsmenlire aqetyqqlsk rlliwlearq | ||
| + | 541 ifdlerslkp htslsisfps dflkkedgls lnndfkkpac nnvtnsdeye qlinkltslq | ||
| + | 601 asrkkdtlyi rklyeelgid dtvnsyrrli amscginped lsleildave ealtrek | ||
| + | " | ||
| + | db <- setDB(database = db, table="protein", id=2, sequence=seq) | ||
| + | |||
| + | </source> | ||
| + | |||
| + | The function to '''add''' a new entry to the database looks quite similar. But we '''have''' to add a test whether the species name has been entered if it's a new taxonomy ID. | ||
| + | |||
| + | <source lang="R"> | ||
| + | |||
| + | addToDB <- function(database, | ||
| + | name = "", | ||
| + | refseq_id = "", | ||
| + | uniprot_id = "", | ||
| + | taxonomy_id = "", | ||
| + | genome_xref = "", | ||
| + | genome_from = "", | ||
| + | genome_to = "", | ||
| + | sequence = "", | ||
| + | species_name = "") { | ||
| + | if (missing(database) | missing(taxonomy_id)) { | ||
| + | stop("database and taxonomy_id have to be specified") | ||
| + | } | ||
| + | # handle taxonomy | ||
| + | if (! any(db$taxonomy$id == taxonomy_id)) { # new taxonomy_id | ||
| + | if (missing(species_name)) { | ||
| + | stop("taxonomy_id is new: species_name has to be specified") | ||
| + | } | ||
| + | else { | ||
| + | # add this species to the taxonomy table | ||
| + | database$taxonomy <- rbind(database$taxonomy, | ||
| + | data.frame(id = taxonomy_id, | ||
| + | species_name = species_name, | ||
| + | stringsAsFactors = FALSE)) | ||
| + | |||
| + | } | ||
| + | } | ||
| + | # handle protein | ||
| + | # get a new protein id | ||
| + | |||
| + | pid <- max(database$protein$id) + 1 | ||
| + | |||
| + | database$protein <- rbind(db$protein, | ||
| + | data.frame(id = pid, | ||
| + | name = name, | ||
| + | refseq_id = refseq_id, | ||
| + | uniprot_id = uniprot_id, | ||
| + | taxonomy_id = taxonomy_id, | ||
| + | genome_xref = genome_xref, | ||
| + | genome_from = genome_from, | ||
| + | genome_to = genome_to, | ||
| + | sequence = sanitizeSequence(sequence), | ||
| + | stringsAsFactors = FALSE)) | ||
| + | |||
| + | return(database) | ||
| + | } | ||
| + | |||
| + | # Test this. | ||
| + | |||
| + | newSeq <- " | ||
| + | 1 msgdktifka tysgvpvyec iinnvavmrr rsddwlnatq ilkvvgldkp qrtrvlerei | ||
| + | 61 qkgihekvqg gygkyqgtwi pldvaielae ryniqgllqp itsyvpsaad spppapkhti | ||
| + | 121 stsnrskkii padpgalgrs rratsietes evigaapnnv segsmspsps dissssrtps | ||
| + | 181 plpadrahpl hanhalagyn grdannhary adiildyfvt enttvpslli npppdfnpdm | ||
| + | 241 sidddehtal hwacamgrir vvklllsaga difrvnsnqq talmratmfs nnydlrkfpe | ||
| + | 301 lfellhrsil nidrndrtvf hhvvdlalsr gkphaaryym etminrlady gdqladilnf | ||
| + | 361 qddegetplt maararskrl vrlllehgad pkirnkegkn aedyiieder frsspsrtgp | ||
| + | 421 agielgadgl pvlptsslht seagqrtagr avtlmsnllh sladsydsei ntaekkltqa | ||
| + | 481 hgllkqiqte iedsakvaea lhheaqgvde erkrvdslql alkhainkra rddlerrwse | ||
| + | 541 gkqaikrarl qaglepgals tsnatnapat gdqkskddak sliealpagt nvktaiaelr | ||
| + | 601 kqlsqvqank telvdkfvar areqgtgrtm aayrrliaag cggiapdevd avvgvlcell | ||
| + | 661 qeshtgarag aggerddrar dvammlkgag aaalaanaga p | ||
| + | " | ||
| + | |||
| + | db2 <- addToDB(db, | ||
| + | name = "UMAG_1122", | ||
| + | refseq_id = "XP_011392621", | ||
| + | uniprot_id = "A0A0D1DP35", | ||
| + | taxonomy_id = "5270", | ||
| + | genome_xref = "NC_026499.1", | ||
| + | genome_from = "150555", | ||
| + | genome_to = "152739", | ||
| + | sequence = newSeq, | ||
| + | species_name = "Ustilago maydis") | ||
| + | |||
| + | </source> | ||
| + | |||
| + | As you can see, adding a new protein to our database is now rather compact. | ||
| + | |||
| + | |||
| + | {{task|1= | ||
| + | |||
| + | In the last assignment, you discovered a protein that is related to yaest Mbp1 in YFO. | ||
| + | |||
| + | Add it to the database. Ask on the mailing list if you don't know how (but be specific) or if you don't know how to find particular information items. | ||
| + | |||
| + | }} | ||
| + | |||
| + | |||
| + | |||
| + | ;TBC | ||
====Storing and Retrieving data==== | ====Storing and Retrieving data==== | ||
Revision as of 12:33, 3 October 2015
Assignment for Week 3
Sequence Analysis
Note! This assignment is currently inactive. Major and minor unannounced changes may be made at any time.
Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz.
Contents
Introduction
One of the foundations of bioinformatics is the empirical observation that related sequences conserve structure, and often function. This is the basis on which we can make inferences from well-studied model organisms in species that have not been studied as deeply. The model case for our assignments is to take annotations from baker's yeast, Saccharomyces cerevisiae and apply them to YFO.
Therefore, in this assignment we will
- use the sequence search program BLAST to retrieve a sequence similar to yeast Mbp1 in YFO;
- use a number of tools to annotate the sequence.
Keeping with our theme of sequence analysis, we will
- explore EMBOSS tools;
- compute and plot relative amino acid frequencies in R;
- and (optionally) use Chimera to explore H-bond patterns in the Mbp1 APSES domain structure.
Retrieve
In Assignment 2 you looked at sequences in YFO that are related to yeast Mbp1, by following a link from the RefSeq record. I mentioned that there are more principled ways to find related proteins: that principle is to search for similar sequences. Exactly how this works will be the subject of later lectures, but the tool that is most commonly used for this task is called BLAST (Basic Local Alignment And Search Tool). The task of this assignment is to perform a number of sequence annotations to the sequence from YFO that is most similar to Mbp1, or, more precisely, that contains an APSES domain that is most similar[1].
Search input
First, we need to define the sequence we will search with, as the search input.
Defining the sequence to search with
I have highlighted the extent of the APSES domain sequence in the previous assignment, but when you explored the corresponding structure in Chimera, you saw that the structured protein domain is larger and the additional secondary structure elements are in fact well integrated into the overall domain. This is not surprising: canonical domain definitions are compiled from many species and examples, and they generally comprise only the common core. Looking up the source of the domain annotations for Mbp1 is very easy:
Task:
- Access the RefSeq record for yeast Mbp1.
- While you are here, download a FASTA formatted version of the sequence to your R working directory and give it a filename of
mbp1-sacce.fa. We will need it later. It should be straightforward from the NCBI page how to achieve that. As a hint, you need to use the Send to... link to actually download the file. - On the RefSeq page, look for the link Related Information → CDD Search Results and follow it.
This is a domain annotation: CDD is the NCBI's Conserved Domain Database and the annotation was done by a tool that scanned the sequence of Mbp1 for segments that are similar to any of the domain definitions stored in the CDD. We will return to CDD in the next assignment.
- Click on the blue box labeled Kila-N in the graph to access the CDD entry for this domain.
- Read the abstract. You should understand the relationship between Kila-N and APSES domains. One is a subfamily of the other.
- Confirm that the domain definition – as applied to the Mbp1 sequence (which is labeled as "query") – corresponds to the region we highlighted in the last assignment.
What precisely constitutes an APSES domain however is a matter of definition, as you can explore in the following (optional) task.
- Display the protein in ribbon style, e.g. with the Interactive 1 preset.
- Access the Interpro information page for Mbp1 at the EBI: http://www.ebi.ac.uk/interpro/protein/P39678
- In the section Domains and repeats, mouse over the red annotations and note down the residue numbers for the annotated domains. Also follow the links to the respective Interpro domain definition pages.
At this point we have definitions for the following regions on the Mbp1 protein ...
- The KilA-N (pfam 04383) domain definition as applied to the Mbp1 protein sequence by CDD;
- The InterPro KilA, N-terminal/APSES-type HTH, DNA-binding (IPR018004) definition annotated on the Mbp1 sequence;
- The InterPro Transcription regulator HTH, APSES-type DNA-binding domain (IPR003163) definition annotated on the Mbp1 sequence;
- (... in addition – without following the source here – the UniProt record for Mbp1 annotates a "HTH APSES-type" domain from residues 5-111)
... each with its distinct and partially overlapping sequence range. Back to Chimera:
- In the sequence window, select the sequence corresponding to the Interpro KilA-N annotation and colour this fragment red. Remember that you can get the sequence numbers of a residue in the sequence window when you hover the pointer over it - but do confirm that the sequence numbering that Chimera displays matches the numbering of the Interpro domain definition.
- Then select the residue range(s) by which the CDD KilA-N definition is larger, and colour that fragment orange.
- Then select the residue range(s) by which the InterPro APSES domain definition is larger, and colour that fragment yellow.
- If the structure contains residues outside these ranges, colour these white.
- Study this in a side-by-side stereo view and get a sense for how the extra sequence beyond the Kil-A N domain(s) is part of the structure, and how the integrity of the folded structure would be affected if these fragments were missing.
- Display Hydrogen bonds, to get a sense of interactions between residues from the differently colored parts. First show the protein as a stick model, with sticks that are thicker than the default to give a better sense of sidechain packing:
- (i) Select → Select all
- (ii) Actions → Ribbon → hide
- (iii) Select → Structure → protein
- (iv) Actions → Atoms/Bonds → show
- (v) Actions → Atoms/Bonds → stick
- (vi) click on the looking glass icon at the bottom right of the graphics window to bring up the inspector window and choose Inspect ... Bond. Change the radius to 0.4.
- (i) Select → Select all
- Then calculate and display the hydrogen bonds:
- (vii) Tools → Surface/Binding Analysis → FindHbond
- (viii) Set the Line width to 3.0, leave all other parameters with their default values an click Apply
- Clear the selection.
- (vii) Tools → Surface/Binding Analysis → FindHbond
- Let's now simplify the scene a bit and focus on backbone/backbone H-bonds:
- (ix) Select → Structure → Backbone → full
- (x) Actions → Atoms/Bonds → show only
- Clear the selection.
- (ix) Select → Structure → Backbone → full
- Save the resulting image as a jpeg no larger than 600px across and upload it to your Lab notebook on the Wiki.
- When you are done, congratulate yourself on having earned a bonus of 10% on the next quiz.
There is a rather important lesson in this: domain definitions may be fluid, and their boundaries may be computationally derived from sequence comparisons across many families, and do not necessarily correspond to individual structures. Make sure you understand this well.
Given this, it seems appropriate to search the sequence database with the sequence of an Mbp1 structure–this being a structured, stable, subdomain of the whole that presumably contains the protein's most unique and specific function. Let us retrieve this sequence. All PDB structures have their sequences stored in the NCBI protein database. They can be accessed simply via the PDB-ID, which serves as an identifier both for the NCBI and the PDB databases. However there is a small catch (isn't there always?). PDB files can contain more than one protein, e.g. if the crystal structure contains a complex[2]. Each of the individual proteins gets a so-called chain ID–a one letter identifier– to identify them uniquely. To find their unique sequence in the database, you need to know the PDB ID as well as the chain ID. If the file contains only a single protein (as in our case), the chain ID is always A[3]. make sure you understand the concept of protein chains, and chain IDs.
Task:
- Back at the RefSeq record for yeast Mbp1, enter the PDB-ID, an underscore, and the chain ID for one of the crystal structures into the search field. You can use
1MB1_Aor1BM8_A, but don't use1L3G: this NMR structure includes a large stretch of unstructured residues. - Click on Display settings and choose FASTA (text). You should get something like:
>gi|157830387|pdb|1BM8|A Chain A, Dna-Binding Domain Of Mbp1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEVLKETHEKVQGGFGKY QGTWVPLNIAKQLAEKFSVYDQLKPLFDF
- Save this sequence in your notebook, in case we need it later.
Next, we use this sequence to find its most similar relative in YFO using BLAST.
BLAST search
Task:
- Navigate to the BLAST entry page at the NCBI.
- Click on protein blast as the BLAST program to run.
- Paste the sequence of the yeast Mbp1 DNA-binding domain into the search field.
- Set the following parameters:
- As Database option choose Reference proteins (refseq_protein)
- As Organism enter the binomial name of YFO. Make sure you spell it right, the page will try to autocomplete your entry. Species level is detailed enough, you don't have to specify the strain (e.g. I would specify "Ustilago maydis" not "Ustilago maydis 521").
- Then click on the BLAST button and wait for the result to appear. You will first see a graph of any conserved domains in your query sequence, this is not yet what you are waiting for...
- Patience.
- Patience. The database is large.
- Patience. Execution times vary greatly by time of day.
- The top "hit" on the results page is what you are looking for. Its alignment and alignment score are shown in the Alignments section a bit further down the page. Your hit should have on the order of more than 40% identities to the query and match at least 80 residues or so. If your match seems less and worse than that, please eMail me to troubleshoot.
- The first item for each hit is a link to its database entry, right next to the checkbox. It says something like
ref|NP_123456789orref|XP_123456789... follow that link. - Note the RefSeq ID, and save the sequence in FASTA format into your R working directory, as you did for Mbp1 at the beginning of the assignment. Give this a filename of
mbp1-xxxxx.fa, but replacexxxxxwith its short species label for YFO. For simplicity I will refer to this sequence as "YFO Mbp1" in the future.
Store
The Protein datamodel
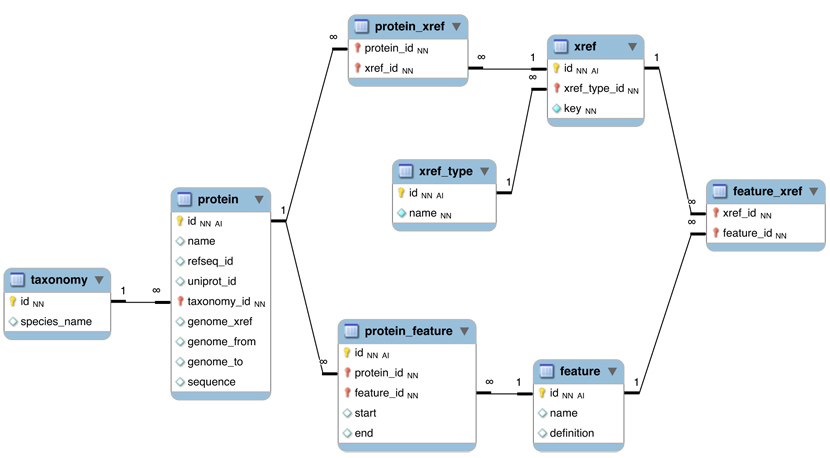
Here is a revised schema for the initial model that we developed in class:
- The protein table is at the centre. Its primary key is a unique integer. We store the NCBI RefSeq ID and the Uniprot ID in the table. We would not call them "foreign keys", since the information they reference is not in our schema but at the NCBI/EBI. The genome_xref field will hold the ID for the actual genomic sequence, where the coding region of the gene is identified by its start ("from") and end ("to").
- The taxonomy table holds information about species. We use the NCBI taxonomy ID as its primary key. The same key appears in the protein table as the foreign key that links the protein with its proper taxonomy information.
- The protein_feature table links a protein with all it's features that we record. Start and end coordinates identify the region of the sequence we have annotated. In principle these attributes could be NULL.
- The feature table contains a feature name and a textual definition. Again, these attributes could theoretically be NULL.
- The protein_xref table and the feature_xref table connect cross-reference data.
- The xref table needs to hold two attributes: the type of cross-reference (e.g. PubMed), from which we would construct an URL or other accession method, and the actual key (e.g. 10747782). The type of cross-reference needs some thought however: this field needs a "controlled vocabulary" to ensure that the same cross-reference is described consistently with the same string (pubmed? PubMed? Pub-Med?). There are a number of ways to achieve this, the best way is to store these types in their own table and link to the table via a foreign key.
Implementing the Data Model in R
To actually implement the model in R we will create the tables as data frames, and we will collect them in a list. We don't have to keep the tables in a list - we could also keep them as independent objects, but with a table named "protein" we should be worried of inadvertently overwriting the table. A list is neater, more flexible (we might want to have several of these), it reflects our intent about the model better, and doesn't require very much more typing.
db <- list()
db$protein <- data.frame(id = 1,
name = "Mbp1",
refseq_id = "NP_010227",
uniprot_id = "P39678",
taxonomy_id = 4932,
genome_xref = "NC_001136.10",
genome_from = 352877,
genome_to = 355378,
sequence = "...",
stringsAsFactors = FALSE)
db$taxonomy <- data.frame(id = 4932,
species_name = "Saccharomyces cerevisiae",
stringsAsFactors = FALSE)
# Let's add a second protein:
db$protein <- rbind(db$protein,
data.frame(id = 2,
name = "Res2",
refseq_id = "NP_593032",
uniprot_id = "P41412",
taxonomy_id = 4896,
genome_xref = "NC_003424.3",
genome_from = 686543,
genome_to = 689179,
sequence = "...",
stringsAsFactors = FALSE))
db$taxonomy <- rbind(db$taxonomy,
data.frame(id = 4896,
species_name = "Schizosaccharomyces pombe",
stringsAsFactors = FALSE))
str(db)
# you can look at the contents of the tables in the usual way:
db$protein
db$protein$refseq_id
db$protein[,"name"]
db$taxonomy
db$taxonomy[1,]
# What is the species name for the protein
# whose name is "Mbp1"?
# Get the taxonomy_id. This is a cell in the
# table: db$protein[<row>, <column>]
# <row> is the row in which the value in
# the name column is Mbp1:
db$protein$name == "Mbp1"
# The <column> is called "taxonomy_id". Simply
# insert these two expressions in the square
# brackets.
db$protein[db$protein$name == "Mbp1", "taxonomy_id"]
# Assign the taxonomy_id value ...
x <- db$protein[db$protein$name == "Mbp1", "taxonomy_id"]
# ... and fetch the species_name value from db$taxonomy
db$taxonomy[db$taxonomy$id == x, "species_name"]This works, but you can appreciate why we don't usually want to type all this this by hand - it's much more convenient, with less potential for error to write the necessary get- and set- functions.
We only entered a placeholder for the sequence field. Sequences come in many different flavours when we copy them from a Webpage: there can be whitespace, carriage returns, numbers, they can be upper-case, lower-case mixed-case ...
What we want in our sequence data field is one string that contains the entire sequence, and nothing but upper-case amino-acid code letters.
We'll need to look at how we work with strings in R, how we identify and work with patterns. This is a great time to introduce regular expressions.
A Brief Introduction to Regular Expressions
- Regular expressions are a concise description language to define patterns for pattern-matching in strings.
Truth be told, many programmers have a love-hate relationship with regular expressions. The syntax of regular expressions is very powerful and expressive, but also terse, not always intuitive, and sometimes hard to understand. I'll introduce you to a few principles here that are quite straightforward and they will probably cover 99% of the cases you will encounter.
Here is our test-case: the sequence of Mbp1, copied from the Genbank Webpage.
1 msnqiysary sgvdvyefih stgsimkrkk ddwvnathil kaanfakakr trilekevlk
61 ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf dftqtdgsas pppapkhhha
121 skvdrkkair sastsaimet krnnkkaeen qfqsskilgn ptaaprkrgr pvgstrgsrr
181 klgvnlqrsq sdmgfprpai pnssisttql psirstmgpq sptlgileee rhdsrqqqpq
241 qnnsaqfkei dledglssdv epsqqlqqvf nqntgfvpqq qssliqtqqt esmatsvsss
301 pslptspgdf adsnpfeerf pgggtspiis miprypvtsr pqtsdindkv nkylsklvdy
361 fisnemksnk slpqvllhpp phsapyidap idpelhtafh wacsmgnlpi aealyeagts
421 irstnsqgqt plmrsslfhn sytrrtfpri fqllhetvfd idsqsqtvih hivkrksttp
481 savyyldvvl skikdfspqy rielllntqd kngdtalhia skngdvvffn tlvkmgaltt
541 isnkegltan eimnqqyeqm miqngtnqhv nssntdlnih vntnnietkn dvnsmvimsp
601 vspsdyityp sqiatnisrn ipnvvnsmkq masiyndlhe qhdneikslq ktlksisktk
661 iqvslktlev lkesskdeng eaqtnddfei lsrlqeqntk klrkrliryk rlikqkleyr
721 qtvllnklie detqattnnt vekdnntler lelaqeltml qlqrknklss lvkkfednak
781 ihkyrriire gtemnieevd ssldvilqtl iannnknkga eqiitisnan sha
//
Task:
Navigate to http://regexpal.com and paste the sequence into the lower box. This site is one of a few online regular expression testers and invaluable when you are developing regular expression patterns.
Lets try some expressions:
- Most characters are matched literally.
- Type "
a" in to the upper box and you will see all "a" characters matched. Then replaceawithq. - Now type "
aa" instead. Thenkrnnkk. Sequences of charcters are also matched literally.
- We can specify more than one character to match if we place it in square brackets.
[lq]matcheslORq.[familyvw]matches hydrophobic amino acids.
- Within square brackets, we can specify "ranges".
[1-5]matches digits from 1 to 5.
- Within square brackets, we can specify characters that should NOT be matched, with the caret,
^. [^0-9]matches everything EXCEPT digits.[^a-z]matches everything that is not a lower-case letter.
One of the R functions that uses regular expressions is the function gsub() that replaces characters that match a "regex" with other characters. That is useful for our purpose: we can
- match to all characters that are NOT a letter, and
- replace them by - nothing: the empty string
"".
This deletes them.
Building on this function, we can write a function to set values in our protein table.
setDB <- function(database,
table,
id,
name = "",
refseq_id = "",
uniprot_id = "",
taxonomy_id = "",
genome_xref = "",
genome_from = "",
genome_to = "",
sequence = "",
species_name = "") {
if (missing(database) | missing(table) | missing(id)) {
stop("database, table and id have to be specified")
}
if (table == "protein") {
row <- which(database$protein$id == id)
if (name != "") { database$protein[row, "name"] <- name }
if (refseq_id != "") { database$protein[row, "refseq_id"] <- refseq_id}
if (uniprot_id != "") { database$protein[row, "uniprot_id"] <- uniprot_id}
if (taxonomy_id != "") { database$protein[row, "taxonomy_id"] <- taxonomy_id}
if (genome_xref != "") { database$protein[row, "genome_xref"] <- genome_xref}
if (genome_from != "") { database$protein[row, "genome_from"] <- genome_from}
if (genome_to != "") { database$protein[row, "genome_to"] <- genome_to}
if (sequence != "") { database$protein[row, "sequence"] <- sanitizeSequence(sequence)}
}
if (table == "taxonomy") {
row <- which(database$taxonomy$id == id)
if (species_name != "") { database$taxonomy[row, "species_name"] <- species_name }
}
return(database)
}
# Note that this code could be significantly improved: we are
# not doing any tests that the values are sane.
# But for now I just want to keep the code simple.
db$protein[1,]
db <- setDB(database = db, table="protein", id=1, name="Armadillo")
db$protein[1,]
db <- setDB(database = db, table="protein", id=1, name="Mbp1")
# With this, we can update our entries to add the sequence.
seq
seq <- "
1 msnqiysary sgvdvyefih stgsimkrkk ddwvnathil kaanfakakr trilekevlk
61 ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf dftqtdgsas pppapkhhha
121 skvdrkkair sastsaimet krnnkkaeen qfqsskilgn ptaaprkrgr pvgstrgsrr
181 klgvnlqrsq sdmgfprpai pnssisttql psirstmgpq sptlgileee rhdsrqqqpq
241 qnnsaqfkei dledglssdv epsqqlqqvf nqntgfvpqq qssliqtqqt esmatsvsss
301 pslptspgdf adsnpfeerf pgggtspiis miprypvtsr pqtsdindkv nkylsklvdy
361 fisnemksnk slpqvllhpp phsapyidap idpelhtafh wacsmgnlpi aealyeagts
421 irstnsqgqt plmrsslfhn sytrrtfpri fqllhetvfd idsqsqtvih hivkrksttp
481 savyyldvvl skikdfspqy rielllntqd kngdtalhia skngdvvffn tlvkmgaltt
541 isnkegltan eimnqqyeqm miqngtnqhv nssntdlnih vntnnietkn dvnsmvimsp
601 vspsdyityp sqiatnisrn ipnvvnsmkq masiyndlhe qhdneikslq ktlksisktk
661 iqvslktlev lkesskdeng eaqtnddfei lsrlqeqntk klrkrliryk rlikqkleyr
721 qtvllnklie detqattnnt vekdnntler lelaqeltml qlqrknklss lvkkfednak
781 ihkyrriire gtemnieevd ssldvilqtl iannnknkga eqiitisnan sha
//
"
db <- setDB(database = db, table="protein", id=1, sequence=seq)
seq <- "
1 maprssavhv avysgvevye cfikgvsvmr rrrdswlnat qilkvadfdk pqrtrvlerq
61 vqigahekvq ggygkyqgtw vpfqrgvdla tkykvdgims pilsldideg kaiapkkkqt
121 kqkkpsvrgr rgrkpsslss stlhsvnekq pnssisptie ssmnkvnlpg aeeqvsatpl
181 paspnallsp ndntikpvee lgmleapldk yeeslldffl hpeegripsf lyspppdfqv
241 nsvidddght slhwacsmgh iemiklllra nadigvcnrl sqtplmrsvi ftnnydcqtf
301 gqvlellqst iyavdtngqs ifhhivqsts tpskvaaaky yldcilekli siqpfenvvr
361 lvnlqdsngd tslliaarng amdcvnslls ynanpsipnr qrrtaseyll eadkkphsll
421 qsnsnashsa fsfsgispai ispscsshaf vkaipsissk fsqlaeeyes qlrekeedli
481 ranrlkqdtl neisrtyqel tflqknnpty sqsmenlire aqetyqqlsk rlliwlearq
541 ifdlerslkp htslsisfps dflkkedgls lnndfkkpac nnvtnsdeye qlinkltslq
601 asrkkdtlyi rklyeelgid dtvnsyrrli amscginped lsleildave ealtrek
"
db <- setDB(database = db, table="protein", id=2, sequence=seq)The function to add a new entry to the database looks quite similar. But we have to add a test whether the species name has been entered if it's a new taxonomy ID.
addToDB <- function(database,
name = "",
refseq_id = "",
uniprot_id = "",
taxonomy_id = "",
genome_xref = "",
genome_from = "",
genome_to = "",
sequence = "",
species_name = "") {
if (missing(database) | missing(taxonomy_id)) {
stop("database and taxonomy_id have to be specified")
}
# handle taxonomy
if (! any(db$taxonomy$id == taxonomy_id)) { # new taxonomy_id
if (missing(species_name)) {
stop("taxonomy_id is new: species_name has to be specified")
}
else {
# add this species to the taxonomy table
database$taxonomy <- rbind(database$taxonomy,
data.frame(id = taxonomy_id,
species_name = species_name,
stringsAsFactors = FALSE))
}
}
# handle protein
# get a new protein id
pid <- max(database$protein$id) + 1
database$protein <- rbind(db$protein,
data.frame(id = pid,
name = name,
refseq_id = refseq_id,
uniprot_id = uniprot_id,
taxonomy_id = taxonomy_id,
genome_xref = genome_xref,
genome_from = genome_from,
genome_to = genome_to,
sequence = sanitizeSequence(sequence),
stringsAsFactors = FALSE))
return(database)
}
# Test this.
newSeq <- "
1 msgdktifka tysgvpvyec iinnvavmrr rsddwlnatq ilkvvgldkp qrtrvlerei
61 qkgihekvqg gygkyqgtwi pldvaielae ryniqgllqp itsyvpsaad spppapkhti
121 stsnrskkii padpgalgrs rratsietes evigaapnnv segsmspsps dissssrtps
181 plpadrahpl hanhalagyn grdannhary adiildyfvt enttvpslli npppdfnpdm
241 sidddehtal hwacamgrir vvklllsaga difrvnsnqq talmratmfs nnydlrkfpe
301 lfellhrsil nidrndrtvf hhvvdlalsr gkphaaryym etminrlady gdqladilnf
361 qddegetplt maararskrl vrlllehgad pkirnkegkn aedyiieder frsspsrtgp
421 agielgadgl pvlptsslht seagqrtagr avtlmsnllh sladsydsei ntaekkltqa
481 hgllkqiqte iedsakvaea lhheaqgvde erkrvdslql alkhainkra rddlerrwse
541 gkqaikrarl qaglepgals tsnatnapat gdqkskddak sliealpagt nvktaiaelr
601 kqlsqvqank telvdkfvar areqgtgrtm aayrrliaag cggiapdevd avvgvlcell
661 qeshtgarag aggerddrar dvammlkgag aaalaanaga p
"
db2 <- addToDB(db,
name = "UMAG_1122",
refseq_id = "XP_011392621",
uniprot_id = "A0A0D1DP35",
taxonomy_id = "5270",
genome_xref = "NC_026499.1",
genome_from = "150555",
genome_to = "152739",
sequence = newSeq,
species_name = "Ustilago maydis")As you can see, adding a new protein to our database is now rather compact.
Task:
In the last assignment, you discovered a protein that is related to yaest Mbp1 in YFO.
Add it to the database. Ask on the mailing list if you don't know how (but be specific) or if you don't know how to find particular information items.
- TBC
Storing and Retrieving data
It is certainly possible to edit any component of our data model directly by changing the elements directly. But that's a really bad idea, since it is far too easy to bring the whole model into an inconsistent state. It's much better to write functions that get and set data elements, not only to keep the data consistent, but also because it is much easier, if the model ever changes, to simply edit the function code, rather than having to repeat every single data entry by hand.
So what would an setProteinData function have to look like? It should
- create a new entry if the requested protein does not exist yet;
- update data if the protein exists
Add your sequence
Annotate
Let us perform a few simple sequence annotations with the EMBOSS package, using the Saccharomyces and YFO Mbp1 sequences. As I noted in class, a large number of simple but fundamental sequence analysis tools are combined in the EMBOSS package. This package can be installed locally on your own machine, or run via a public Web interface[4].
Access one of the EMBOSS Explorer services and explore some of the tools:
Task:
- Local composition
- Find
pepinfounder the PROTEIN COMPOSITION heading. - Paste the YFO Mbp1 sequence in FASTA format into the input field.
- Run with default parameters.
- Do the same in a separate window for yeast Mbp1.
- Try to compare ... kind of hard without reference, overlay and expectation, isn't it?
Task:
- Global composition
- Find
pepstatsunder the PROTEIN COMPOSITION heading. - Paste the YFO Mbp1 sequence in FASTA format into the input field.
- Run with default parameters.
- Do the same in a separate window for yeast Mbp1.
- Try to compare ... are there significant and unexpected differences?
Task:
- Motifs
- Find
patmatmotifsandpepcoils. - Run these with the YFO Mbp1 sequence and yeast Mbp1.
- Try to compare ... do both sequences have the same motif annotated in approximately comparable regions of the protein?
Task:
- Transmembrane sequences
- Find
tmap. Also findshuffleseq. - Use your YFO Mbp1 sequence to annotate transmembrane helices for it and for a few shuffled sequences. YFO Mbp1 should have neither, and the shuffled sequences work as negative controls in any case.
- Also compare the following positive contros:
Gef1 - a yeast chloride channel with 10 trans-membrane helices and outward localized N-terminus:
>gi|6322500|ref|NP_012574.1| Gef1p [Saccharomyces cerevisiae S288c]
MPTTYVPINQPIGDGEDVIDTNRFTNIPETQNFDQFVTIDKIAEENRPLSVDSDREFLNSKYRHYREVIW
DRAKTFITLSSTAIVIGCIAGFLQVFTETLVNWKTGHCQRNWLLNKSFCCNGVVNEVTSTSNLLLKRQEF
ECEAQGLWIAWKGHVSPFIIFMLLSVLFALISTLLVKYVAPMATGSGISEIKVWVSGFEYNKEFLGFLTL
VIKSVALPLAISSGLSVGKEGPSVHYATCCGYLLTKWLLRDTLTYSSQYEYITAASGAGVAVAFGAPIGG
VLFGLEEIASANRFNSSTLWKSYYVALVAITTLKYIDPFRNGRVILFNVTYDRDWKVQEIPIFIALGIFG
GLYGKYISKWNINFIHFRKMYLSSWPVQEVLFLATLTALISYFNEFLKLDMTESMGILFHECVKNDNTST
FSHRLCQLDENTHAFEFLKIFTSLCFATVIRALLVVVSYGARVPAGIFVPSMAVGATFGRAVSLLVERFI
SGPSVITPGAYAFLGAAATLSGITNLTLTVVVIMFELTGAFMYIIPLMIVVAITRIILSTSGISGGIADQ
MIMVNGFPYLEDEQDEEEEETLEKYTAEQLMSSKLITINETIYLSELESLLYDSASEYSVHGFPITKDED
KFEKEKRCIGYVLKRHLASKIMMQSVNSTKAQTTLVYFNKSNEELGHRENCIGFKDIMNESPISVKKAVP
VTLLFRMFKELGCKTIIVEESGILKGLVTAKDILRFKRIKYREVHGAKFTYNEALDRRCWSVIHFIIKRF
TTNRNGNVI- Evaluate the output: does the algorithm (wrongly) predict tm-helices in your protein? In the shuffled sequences? Does it find the tm-helices in Gef1?
Try to familiarize yourself with the offerings in the EMBOSS package. I find some of the nucleic acid tools indispensable in the lab, such as restriction-site mapping tools, and I frequently use the alignment tools Needle and Water, but by and large the utility of many of the components–while fast, efficient and straightforward to use– suffers from lack of reference and comparison and from terse output. The routines show their conceptual origin in the 1970s and 1980s. We will encounter alternatives in later assignments.
Plotting sequence composition in R
It is rather straightforward to code a simple amino acid composition measurement in R and I have shown you an example in the lecture.
Task:
- Open R, access the R tutorial and work through the sections on
- Scripts;
- Variables;
- Scalar data; and
- Vectors. This will help you understand the following code.
- Study the following code. Ask on the list if there are parts of the syntax that you don't understand:
# aaFreq.R
# plot amino-acid frequency excess
# Boris Steipe for BCH441, 2012 - 2014
# Go to your R working directory that contains the FASTA
setwd("my/BCH441/R/directory")
# install the package "seqinr"
install.packages("seqinr")
# load the library for seqinr
library(seqinr)
# define the filename for the YFO Mbp1 sequence
seqfile <- "mbp1-schpo.fa"
# load a fasta-formatted sequence from your working directory.
mbp1 <- read.fasta(seqfile, seqtype="AA", set.att=FALSE)
# define reference composition, a database average
# (data taken from http://web.expasy.org/docs/relnotes/relstat.html)
refData <- c(
"A"=8.26,
"Q"=3.93,
"L"=9.66,
"S"=6.56,
"R"=5.53,
"E"=6.75,
"K"=5.84,
"T"=5.34,
"N"=4.06,
"G"=7.08,
"M"=2.42,
"W"=1.08,
"D"=5.45,
"H"=2.27,
"F"=3.86,
"Y"=2.92,
"C"=1.37,
"I"=5.96,
"P"=4.70,
"V"=6.87
)
refData = refData / sum(refData) #Normalize
# how many different characters are in the dataset?
lref <- length(refData)
# tabulate the sequence of interest and normalize
obsData <- table(mbp1) # count occurrences
obsData = obsData / sum(obsData) # Normalize
# initialize a vector of zeros, which will hold our results
logRatio <- rep(lref, 0)
# loop over all elements of the reference and calculate log-ratios
for (i in 1:lref) {
aa <- names(refData)[i] # get the name of that amino acid
fObs <- obsData[aa] # retrieve the frequency for that name
fRef <- refData[aa]
logRatio[aa] <- log(fObs / fRef) / log(2)
}
logRatio <- sort(logRatio, decreasing = TRUE) # sort values descending
# generate a y-axis label that also contains a subscript
# (see text() for details)
label <- expression(paste(log[2],"( f(obs) / f(ref) )", sep = ""))
# define colors for the bars
chargePlus <- "#282A3F"
chargeMinus <- "#531523"
hydrophilic <- "#47707F"
hydrophobic <- "#6F7A79"
plain <- "#EDF7F7"
# Assign the colors to the different amino acid types
barColors <- rep(length(refData), 0)
for (i in 1:length(refData)) {
if (names(logRatio[i]) == "A") { barColors[i] <- hydrophobic }
else if (names(logRatio[i]) == "C") { barColors[i] <- plain }
else if (names(logRatio[i]) == "D") { barColors[i] <- chargeMinus }
else if (names(logRatio[i]) == "E") { barColors[i] <- chargeMinus }
else if (names(logRatio[i]) == "F") { barColors[i] <- hydrophobic }
else if (names(logRatio[i]) == "G") { barColors[i] <- plain }
else if (names(logRatio[i]) == "H") { barColors[i] <- chargePlus }
else if (names(logRatio[i]) == "I") { barColors[i] <- hydrophobic }
else if (names(logRatio[i]) == "K") { barColors[i] <- chargePlus }
else if (names(logRatio[i]) == "L") { barColors[i] <- hydrophobic }
else if (names(logRatio[i]) == "M") { barColors[i] <- hydrophobic }
else if (names(logRatio[i]) == "N") { barColors[i] <- hydrophilic }
else if (names(logRatio[i]) == "P") { barColors[i] <- plain }
else if (names(logRatio[i]) == "Q") { barColors[i] <- hydrophilic }
else if (names(logRatio[i]) == "R") { barColors[i] <- chargePlus }
else if (names(logRatio[i]) == "S") { barColors[i] <- hydrophilic }
else if (names(logRatio[i]) == "T") { barColors[i] <- hydrophilic }
else if (names(logRatio[i]) == "V") { barColors[i] <- hydrophobic }
else if (names(logRatio[i]) == "W") { barColors[i] <- hydrophobic }
else if (names(logRatio[i]) == "Y") { barColors[i] <- hydrophobic }
else { barColors[i] <- plain }
}
# create a plot
barplot(logRatio,
ylim=c(-4,1),
main = paste("AA frequencies for ", seqfile),
col = barColors,
ylab = label,
cex.names=0.9)
# draw a line at 0
abline(h=0)
# create a legend that indicates what the colors mean
legend (x = 1,
y = -1,
legend = c("charged (+)",
"charged (-)",
"hydrophilic",
"hydrophobic",
"plain"),
bty = "n",
fill = c(chargePlus,
chargeMinus,
hydrophilic,
hydrophobic,
plain)
)
- 3. Copy the code and execute it in R. You need the appropriate FASTA file for YFO in your working directory. The yeast Mbp1 protein is enriched in hydrophilic residues and has a slight excess of (+) charged residues. This might be compatible with a partially natively disordered protein with DNA-binding properties. How is YFO Mbp1 similar or different? Make a second plot and compare. The yeast sequence is here. Would you expect the two sequences to have similar amino acid frequencies? Why or why not?
Chimera "sequence": implicit or explicit ?
We discussed the distinction between implicit and explicit sequence. But which one does the Chimera sequence window display? Let's find out.
Task:
- Open Chimera and load the 1BM8 structure from the PDB.
- Save the ccordinate file with File → Save PDB ..., use a filename of
test.pdb. - Open this file in a plain text editor: notepad, TextEdit, nano or the like, but not MS Word! Make sure you view the file in a fixed-width font, not proportionally spaced, i.e. Courier, not Arial. Otherwise the columns in the file won't line up.
- Find the records that begin with
SEQRES ...and confirm that the first amino acid isGLN. - Now scroll down to the
ATOMsection of the file. Identify the records for the first residue in the structure. Delete all lines for side-chain atoms except for theCBatom. This changes the coordinates for glutamine to those of alanine. - Replace the
GLNresidue name withALA(uppercase). This relabels the residue as Alanine in the coordinate section. Therefore you have changed the implicit sequence. Implicit and explicit sequence are now different. The second atom record should now look like this:
ATOM 2 CA ALA A 4 -0.575 5.127 16.398 1.00 51.22 C
- Save the file and load it in Chimera.
- Open the sequence window: does it display
QorAas the first reside?
Therefore, does Chimera use the implicit or explicit sequence in the sequence window?
- That is all.
Links and resources
Footnotes and references
- ↑ As you will see later on in the assignment, Mbp1-related proteins contain "Ankyrin" domains, a very widely distributed protein-protein interaction motif that may give rise to false-positive similarities for full-length sequence searches. Therefore, we search only with the DNA binding domain sequence, since this is the functionality that best characterizes the "function" of the protein we are interested in.
- ↑ Think of the ribosome or DNA-polymerase as extreme examples.
- ↑ Otherwise, you need to study the PDB Web page for the structure, or the text in the PDB file itself, to identify which part of the complex is labeled with which chain ID. For example, immunoglobulin structures some time label the light- and heavy chain fragments as "L" and "H", and sometimes as "A" and "B"–there are no fixed rules. You can also load the structure in VMD, color "by chain" and use the mouse to click on residues in each chain to identify it.
- ↑ Google for EMBOSS explorer, public access points include http://emboss.bioinformatics.nl/ and http://bioinfo.unice.fr/emboss/index.html
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.