Difference between revisions of "Database Exam Questions"
(→2005) |
m |
||
| Line 1: | Line 1: | ||
| + | <div id="BIO"> | ||
| + | <div class="b1"> | ||
| + | Databases Exam Questions | ||
| + | </div> | ||
| + | |||
| + | | ||
| + | | ||
| + | |||
__NOTOC__ | __NOTOC__ | ||
| | ||
| Line 176: | Line 184: | ||
--> | --> | ||
| + | |||
| + | [[Category: Bioinformatics]] | ||
| + | </div> | ||
Latest revision as of 01:56, 11 December 2012
Databases Exam Questions
- One aspect of Bioinformatics concerns itself with the storage, organisation, and retreival of biological information. The questions in this section consider the contents and use of some of the key abstractions (sequences, structures, graphs ...) that we deal with, and the databases we store them in.
2003
| In the excerpt from the PDBsum database shown here, please comment briefly on the following points. Questions correspond to the numbers shown on the image. The region within the circle is an enlargement of the original, for better legibility.
|
2004 - PDB Format
Despite its many shortcomings and inconsistencies, the PDB format for coordinate datasets is still the most widely accepted format, chiefly due to the large number of legacy programs that use it, but also because it is human readable. The following is an excerpt from the PDB file of pea defensin (1JKZ.PDB).
[...] ATOM 404 N ALA A 28 1.084 7.614 2.493 1.00 0.00 N ATOM 405 CA ALA A 28 0.164 7.660 3.616 1.00 0.00 C ATOM 406 C ALA A 28 0.842 7.090 4.856 1.00 0.00 C ATOM 407 O ALA A 28 0.731 5.902 5.139 1.00 0.00 O ATOM 408 CB ALA A 28 -1.123 6.911 3.287 1.00 0.00 C ATOM 409 H ALA A 28 1.535 6.768 2.288 1.00 0.00 H ATOM 410 HA ALA A 28 -0.085 8.696 3.802 1.00 0.00 H ATOM 411 1HB ALA A 28 -1.278 6.918 2.218 1.00 0.00 H ATOM 412 2HB ALA A 28 -1.957 7.396 3.773 1.00 0.00 H ATOM 413 3HB ALA A 28 -1.047 5.891 3.634 1.00 0.00 H [...]
- Which atom numbers correspond to the backbone atoms and which atomnumbers correspond to the sidechain of this aminoacid ?
- Describe the information in the following columns (indicated by the values in the first record):
" N ", "A", "1.084", "1.00", "0.00". - Are any of these columns optional, and if yes, would their absence shift the positions of the other columns?
- Briefly discuss the relationship between
SEQRESrecords in a PDB file, the genetic sequence of a protein, and the sequence that can be derived from the coordinate records.
Here is a sequence file for this protein.
>gi|20139322|sp|P81929|PSD1_PEA Defense-related peptide 1 KTCEHLADTYRGVCFTNASCDDHCKNKAHLISGTCHNWKCFCTQNC
- What is the name of this file-format?
- Find the amino acid for which the coordinates were given above, in this sequence. (Write its one letter code into your exam booklet together with the preceeding and the following amino acid and underline it e.g. ABC ).
In the coordinate file of the immunoglobulin domain 2IMM.pdb you find the following record.
HETATM 877 O HOH 1 -4.169 60.050 40.145 1.00 3.00 O
- What does this record describe ?
- When you display the structure of 2IMM.pdb with RasMol, the protein is displayed as a wireframe model but you see nothing that corresponds to the above record. What do you need to do ?
(Indeed, since the RasMol tutorial was a task of the first assignment, a question like this may turn up every now and then.)
2003 - Entrez
Briefly discuss each database that the following terms link to, what relationship the results have to the search term and what use one can make of the links.
- PubMed
- Protein
- CDD
- OMIM
(A similar question was given in the 2004 practice exam and I like the following format much better:)
Discuss briefly which of the links you would follow to solve the following problems and summarize what the respective database contains and how you would use it. (Be reasonably complete, more than one link may be needed or helpful. Assume you know nothing about the problem but what is stated in the question.)
- Retrieve 1 kb of upstream sequence for each yeast protein that contains an SH3 domain.
- Check whether a mutation in a residue of yeast protein is known to cause disease in its closest human homologue.
2005
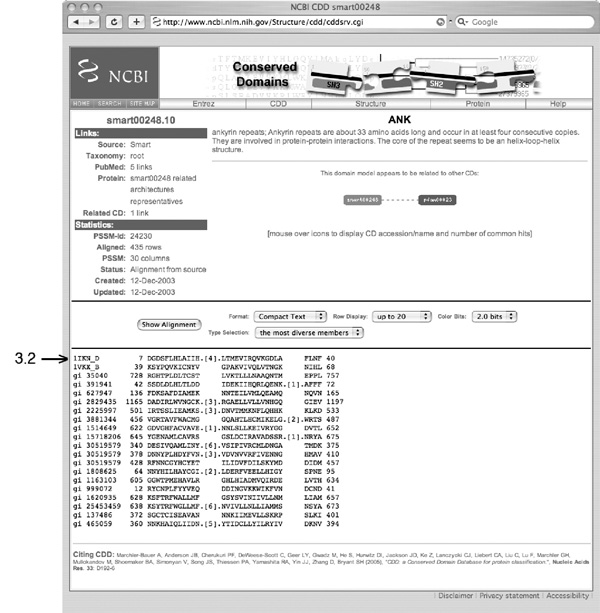
Mbp1 contains ankyrin repeats, these are common protein-protein interaction modules.
- Where does the information that is presented here come from?
- Explain the semantics (meaning) of the identifier that is indicated with the label 3.2
1IKN_D. - Describe one way to identify the source organism of the protein in the third row of the alignment.
- Explain what the numbers in square brackets mean (".[n].") and why they are absent in some rows.
- Describe how the nucleotide sequence for the protein in the third row of the alignment can be retrieved.
2006 Yeast Genome Browser
The summary paragraph for Mbp1 on the record at the Saccharomyces Genome Database (SGD) states:
- Mbp1p is a DNA-binding protein that forms MBF complex (Mlu1 cell cycle box [MCB] Binding Factor) with Swi6p. MBF is a sequence-specific transcription factor that regulates gene expression during the G1/S transition of the cell cycle. Several genes activated or repressed by MBF have been identified, many of which are involved in DNA synthesis and DNA repair (for example, CDC21, CDC8, and CDC9, and also G1 cyclins).
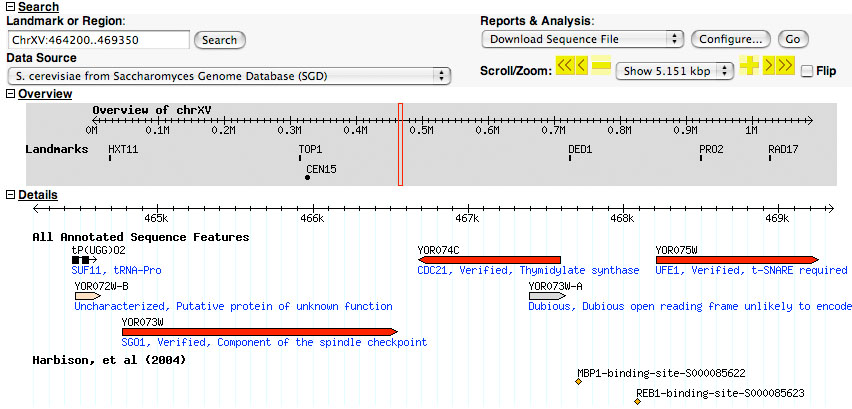
The record for CDC21 links to the following genome-browser page:
- Which genomic region is being displayed?
- What is the signifcance of the arrows? Why do they go in different directions?
- What makes YOR073W-A particularly "dubious"?
- Where is the information that Cdc21 is regulated by Mbp1 shown?
- The label "Harbison et al (2004)" is a so called "Track"; such Tracks can be switched on or off to customize the information that is being presented. What information does this particular track contain?
This question related to biological facts that were known in principle from course assignments, but this particular view and the information that is contained in such genome browser pages was not spelled out explicitly. I expect that this would have been new to most students. One of the course's objectives is to teach how to use novel services and results: analyze what you see, apply your background knowledge, interpret the meaning of the presentation.