Difference between revisions of "Homology modeling fallback data"
Jump to navigation
Jump to search
m (→Model data) |
|||
| (11 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | <div id="BIO"> | |
| − | <div style=" | + | <div class="b1"> |
| − | + | Homology Modeling<br /> | |
| + | <span style="font-size: 70%">Fallback Data</span> | ||
</div> | </div> | ||
| + | |||
| + | |||
| + | Here are results from a homology modeling exercise using SwissModel, as a '''fallback for technical problems with the assignment''' i.e network problems or other issues with the program or input data. | ||
| + | |||
| | ||
| | ||
| Line 10: | Line 15: | ||
==Target sequence== | ==Target sequence== | ||
| − | > | + | This is just one of the reference species' sequences - I chose <code>Mbp1_SCHPO</code> because it is the most distantly related to ''saccharomyces cerevisiae''. |
| − | + | ||
| − | + | >MBP1_SCHPO NP_593032 23..96 | |
| + | IKGVSVMRRRRDSWLNATQILKVADFDKPQRTRVLERQVQIGAHEKVQGGYGKYQGTWVPFQRGVDLATK | ||
| + | YKV | ||
| + | |||
==FASTA formatted target-template alignment== | ==FASTA formatted target-template alignment== | ||
| − | > | + | This uses the 1BM8 PDB file as template. |
| − | + | ||
| − | + | >1BM8_A | |
| − | > | + | QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI |
| − | + | LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF | |
| − | + | >Mbp1_SCHPO 2-100 NP_593032 | |
| + | AVHVAVYSGVEVYECFIKGVSVMRRRRDSWLNATQILKVADFDKPQRTRV | ||
| + | LERQVQIGAHEKVQGGYGKYQGTWVPFQRGVDLATKYKVDGIMSPILSL | ||
| + | |||
| + | There are no gaps and the sequences align over the whole length. If the sequences were of different length, the shorter one would have to be padded with gap characters: <code>"-"</code>. | ||
==SwissModel response== | ==SwissModel response== | ||
| − | |||
| − | = | + | Explanation of the output is found [http://swissmodel.expasy.org/workspace/index.php?func=special_help '''in the SwissModel Help page''']. |
| + | |||
| + | [[Image:ModelledRange.png|frame|none|Graphical comparison of template and target sequence to emphasize which regions have been modelled. Given the good, full-length alignment, SwissModel had no problem modeling the entire protein.]] | ||
| + | |||
| + | |||
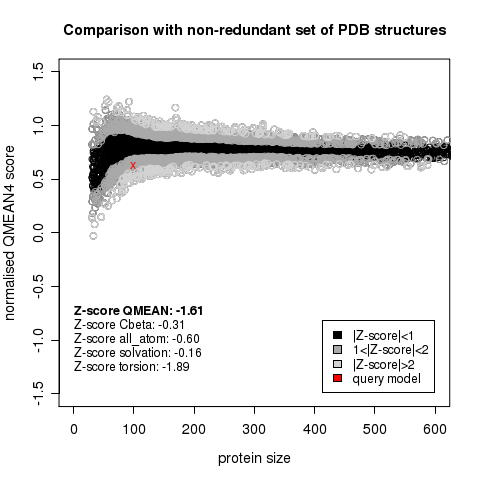
| − | * [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/ | + | [[Image:QMeanScore.png|frame|none|Model QMEAN score vs. expected QMean scores of protein structures, as a function of domain size. The model (red cross) is within the lower 2 sigma range in quality for its size.]] |
| + | |||
| + | |||
| + | |||
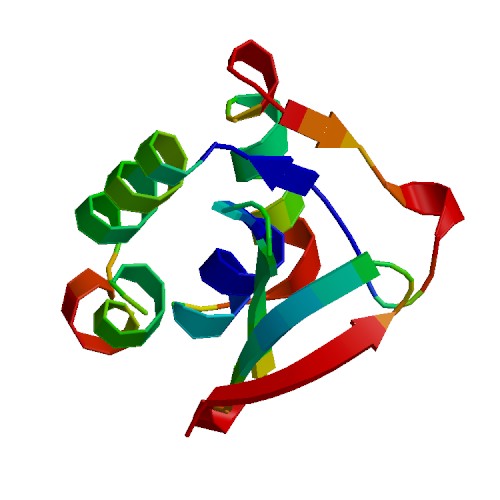
| + | [[Image:ColoringByScore.jpg|frame|none|Cartoon view of the model, colored by QMEAN scores]] | ||
| + | |||
| + | |||
| + | |||
| + | The target-template alignment: | ||
| + | |||
| + | TARGET 1 AVHVAVYS GVEVYECFIK GVSVMRRRRD SWLNATQILK VADFDKPQRT | ||
| + | 1bm8A 4 qiysarys gvdvyefihs tgsimkrkkd dwvnathilk aanfakakrt | ||
| + | |||
| + | TARGET sssssss ssssssss sssssss ssshhhhh h hhhhh | ||
| + | 1bm8A sssssss ssssssss sssssss ssshhhhh h hhhhh | ||
| + | |||
| + | |||
| + | TARGET 49 RVLERQVQIG AHEKVQGGYG KYQGTWVPFQ RGVDLATKYK VDGIMSPILS | ||
| + | 1bm8A 52 rilekevlke thekvqggfg kyqgtwvpln iakqlaekfs vydqlkplfd | ||
| + | |||
| + | TARGET hhhhhh sss ssss hh hhhhhhhh hh hhhh | ||
| + | 1bm8A hhhhhh sss ssss hh hhhhhhhh hh hhhh | ||
| + | |||
| + | |||
| + | TARGET 99 L | ||
| + | 1bm8A 102 f- | ||
| + | |||
| + | TARGET | ||
| + | 1bm8A | ||
| + | |||
| + | |||
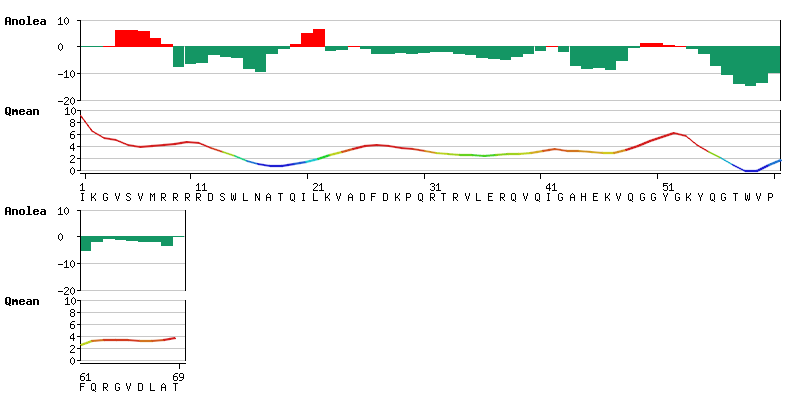
| + | [[Image:ModelQualityEstimation.png|frame|none|Local quality estimation: '''ANOLEA''' and '''QMEAN''' scores as graphical output]] | ||
| + | |||
| + | ==Model data== | ||
| + | |||
| + | * [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/resources/SCHPO_model.pdb SCHPO_model.pdb]: MBP1_SCHPO APSES domain model coordinates | ||
| + | * [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/resources/Local_energy_profile.csv Local_energy_profile.csv]: Energy scores for the MBP1_SCHPO APSES domain model coordinates | ||
| + | |||
| + | |||
| + | | ||
| + | [[Category:Bioinformatics]] | ||
| + | </div> | ||
Latest revision as of 02:41, 5 November 2012
Homology Modeling
Fallback Data
Here are results from a homology modeling exercise using SwissModel, as a fallback for technical problems with the assignment i.e network problems or other issues with the program or input data.
Contents
Target sequence
This is just one of the reference species' sequences - I chose Mbp1_SCHPO because it is the most distantly related to saccharomyces cerevisiae.
>MBP1_SCHPO NP_593032 23..96 IKGVSVMRRRRDSWLNATQILKVADFDKPQRTRVLERQVQIGAHEKVQGGYGKYQGTWVPFQRGVDLATK YKV
FASTA formatted target-template alignment
This uses the 1BM8 PDB file as template.
>1BM8_A QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF >Mbp1_SCHPO 2-100 NP_593032 AVHVAVYSGVEVYECFIKGVSVMRRRRDSWLNATQILKVADFDKPQRTRV LERQVQIGAHEKVQGGYGKYQGTWVPFQRGVDLATKYKVDGIMSPILSL
There are no gaps and the sequences align over the whole length. If the sequences were of different length, the shorter one would have to be padded with gap characters: "-".
SwissModel response
Explanation of the output is found in the SwissModel Help page.
The target-template alignment:
TARGET 1 AVHVAVYS GVEVYECFIK GVSVMRRRRD SWLNATQILK VADFDKPQRT
1bm8A 4 qiysarys gvdvyefihs tgsimkrkkd dwvnathilk aanfakakrt
TARGET sssssss ssssssss sssssss ssshhhhh h hhhhh
1bm8A sssssss ssssssss sssssss ssshhhhh h hhhhh
TARGET 49 RVLERQVQIG AHEKVQGGYG KYQGTWVPFQ RGVDLATKYK VDGIMSPILS
1bm8A 52 rilekevlke thekvqggfg kyqgtwvpln iakqlaekfs vydqlkplfd
TARGET hhhhhh sss ssss hh hhhhhhhh hh hhhh
1bm8A hhhhhh sss ssss hh hhhhhhhh hh hhhh
TARGET 99 L
1bm8A 102 f-
TARGET
1bm8A
Model data
- SCHPO_model.pdb: MBP1_SCHPO APSES domain model coordinates
- Local_energy_profile.csv: Energy scores for the MBP1_SCHPO APSES domain model coordinates