|
|
| Line 2: |
Line 2: |
| | {{Template:Active}} | | {{Template:Active}} |
| | | | |
| − | <br>
| + | |
| | | | |
| | __TOC__ | | __TOC__ |
| − | | + | |
| − | <br> | + | |
| | | | |
| | <div style="padding: 5px; background: #A6AFD0; border:solid 1px #AAAAAA; font-size:200%;font-weight:bold;"> | | <div style="padding: 5px; background: #A6AFD0; border:solid 1px #AAAAAA; font-size:200%;font-weight:bold;"> |
| − | Assignment 3 - Multiple Sequence Alignment | + | Assignment 4 - Phylogenetic Analysis |
| | </div> | | </div> |
| − |
| |
| − | <br>
| |
| − |
| |
| − | {{Template:Preparation|
| |
| − | care=Be sure you have understood all parts of the assignment and cover all questions in your answers! Sadly, we always get assignments back in which people have simply overlooked crucial questions. Sadly, we always get assignments back in which people have not described procedural details. If you did not notice that the above were two different sentences, you are still not reading carefully enough.|
| |
| − | num=3|
| |
| − | ord=third|
| |
| − | due = Monday, November 21. at 12:00}}
| |
| − |
| |
| − | ;Your documentation for the procedures you follow in this assignment will be worth 1 mark.
| |
| − |
| |
| − |
| |
| − | <br>
| |
| | | | |
| | <div style="padding: 2px; background: #F0F1F7; border:solid 1px #AAAAAA; font-size:125%;color:#444444"> | | <div style="padding: 2px; background: #F0F1F7; border:solid 1px #AAAAAA; font-size:125%;color:#444444"> |
| | Introduction | | Introduction |
| | + | |
| | | | |
| − | <br>
| + | ;Nothing in Biology makes sense except in the light of evolution. |
| − | | + | :''Theodosius Dobzhansky'' |
| − | ;Take care of things, and they will take care of you.
| |
| − | :''Shunryu Suzuki'' | |
| | </div> | | </div> |
| | | | |
| − | Much of what we know about a protein's physiological function is based on the '''conservation''' of that function as the species evolves. We assess conservation by comparison to related proteins. Conservation - or variability - is a consequence of '''selection under constraints''': the multiple effects on a species' fitness function that are induced through changes to the structural or functional features of a protein. Conservation patterns can thus provide evidence for many different questions: structural conservation among proteins with similar 3D-structures, functional conservation among homologues with comparable roles, peaks of sequence variability that indicate domain boundaries in multi-domain proteins, or amino acid propensities as predictors for protein engineering and design tasks.
| + | ... but does evolution make sense in the light of biology? |
| | | | |
| − | Measuring conservation requires alignment. Therefore a carefully done multiple sequence alignment (MSA) is a cornerstone for the annotation of the essential properties a gene or protein. MSAs are also useful to resolve ambiguities in the precise placement of indels and to ensure that columns in alignments actually contain amino acids that evolve in a similar context. MSAs serve as input for
| + | As we have seen in the previous assignments, the Mbp1 transcription factor has homologues in all other fungi, yet - looking at orthologues - this is not always a clear one-to-one mapping of related genes to each other. It appears that various systems of APSES domain transcription factors have evolved independently. Of course this bears directly on our notion of ''function'' - what it means to say that two genes in different organisms have the "same" function. In case two organisms both have an orthologous gene for the same, distinct function, this may be warranted. But what if that gene has duplicated in one of them, and the two paralogues now perform different, related functions in one organism? In order to be able to even ask such questions, we need to understand how we can make the evolutionary history of gene families explicit. This is the domain of '''phylogenetic analysis'''. We can ask questions like: how many paralogues did the cenancestor of a clade possess? Which of these underwent additional duplications in the phylogenesis of the organism I am studying? Did any genes get lost? And - adding additional biological insight to the picture - did the observed duplications lead to the "invention" of new biological systems? When was that? And how did the species benefit from this event? |
| − | * functional annotation;
| |
| − | * protein homology modeling;
| |
| − | * phylogenetic analyses, and
| |
| − | * sensitive homology searches in databases.
| |
| | | | |
| | + | We will develop this kind of analysis in this assignment. In the previous assignment you have established which genes are the reciprocally most closely related orthologues to Mbp1 and to other yeast APSES domain genes. In this assignment, we will analyse their evolutionary relationship and compare it to the evolutionary relationship of all fungal APSES domains. The goal is to define families of related transcription factors and their evolutionary history. |
| | | | |
| − | As a first step, we will explore the search and retrieval of fungal proteins that are orthologous to yeast Mbp1, and of the APSES domains they contain. Each student is being assigned one genome-sequenced fungus. Briefly, you will
| + | A number of good tools for phylogenetic analysis exist; ''general purpose packages'' include the (free) [http://evolution.genetics.washington.edu/phylip.html PHYLIP] package and the (commercial) PAUP package. ''Specialized tools'' for tree-building include Treepuzzle or Mr. Bayes. This assignment is conctructed around programs that are availble in PHYLIP, however you are welcome to use other tools that fulfil a similar purpose if you wish. In this field, researchers consider trees that have been built with ML (maximum likelihood) methods to be more reliable than trees that are built with parsimony methods, or distance methods such as NJ (Neighbor Joining). However ML methods are also much more compute-intensive. Just like with multiple sequence alignments, some algorithms will come closer to guessing the truth and others will not and usually it is hard to tell which is the more trustworthy of two diverging results. The prudent researcher tries out alternatives and forms her own opinion. Specifically, we may usually assume results that converge, independent of the algorithm, to be more reliable than those that depend strongly on a particular algorithm or details of input data. |
| | | | |
| − | # Collect sequence identifiers for all APSES domain transcription factors in [[Species list|your assigned species]];
| + | But regarding algorithm and rersources: we will take two shortcuts in this assignment (and both shortcuts are things you should not do ''in real life''): |
| − | # Retrieve the sequences;
| |
| − | # Perform a multiple sequence alignment with these, and a number of reference domains;
| |
| − | # Edit the alignment and annotate.
| |
| | | | |
| | + | '''One''': we will use an '''efficient''' tree-building algorithm, not the best-available one. This is an algorithm which is available through an online Webserver, without the need for you to install software on your own machine. In ''real life'' you would of course use the most accurate algortihm you can get, regardless of the resources this requires, since it makes no sense to waste your time on a careful analysis of inaccurate trees. Your supervisor would want it so as well. And if not she, the reviewers of your manuscript. <small>(However, the simpler algorithm we use here apears to give results that appear quite plausible for the situation we are studying.)</small> |
| | | | |
| − | Multiple Sequence Alignment is not a solved, computational problem and a significant number of alignment tools exist, each with different strengths and objectives. It is remarkable that by far the most frequently used MSA algorithm is CLUSTAL, a procedure that was first published for the microprocessors of the late 1980s, surpassed in performance many times, and shown to be significantly inferior to more modern approaches when aligning sequences with 30% identity or less. In this assignment we will encounter various approaches to multiple alignment:
| + | '''Two''': we will assume the tree the algorithm constructs is ''correct''. In ''real life'' you would establish its reliability with a bootstrap procedure: repeat the tree-building a hundred times with partial data and see which branches and groupings are robust and which depend on the details of the data. However, we should acknowledge that bifurcations that are very close to each other have not been" resolved". Any conscientious reviewer would flag such leniency and send your results back to you for a bootstrapping exercise at the computer. In phylogenetic analysis, not all lines a program draws are equally trustworthy. Dont take the trees as a given fact just because a program suggests this. Look at the evidence, include independent information where available, use your reasoning, and analyse the reults critically. |
| | | | |
| − | * A model-based approach (based on the [[Glossary#PSSM| PSSM]] that PSI-BLAST generates)
| + | In case you want to review concept of trees, clades, LCAs OTUs and the like, I have linked an excellent and very understandable introduction-level [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/restricted/Baldauf_2003_PhylogenyTutorial.pdf article on phylogenetic analysis (pdf)] here and to the resource section at the bottom of this page. |
| − | * Progressive alignments - CLUSTAL and MAFFT
| |
| − | * Consistency based alignment - T-Coffee and MUSCLE
| |
| | | | |
| | + | |
| | | | |
| − | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;">
| + | {{Template:Preparation| |
| − | ==(1) Mbp1 homologues== | + | care=Be sure you have understood all parts of the assignment and cover all questions in your answers! Sadly, we always get assignments back in which important aspects have simply overlooked marks unnecessarily. If you did not notice that the above did not make sense, you are reading what you expect, not what is written.| |
| − | </div>
| + | num=4| |
| | + | ord=fourth| |
| | + | due = Monday, November 17 at 10:00 in the morning}} |
| | | | |
| | + | ;Your documentation for the procedures you follow in this assignment will be worth 1 mark. |
| | | | |
| − | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;">
| + | |
| − | ===(1.1) Retrieving sequences===
| + | |
| − | </div>
| |
| | | | |
| − | | + | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> |
| − | In [[Assignment 2]] you retrieved the protein sequences of ''saccharomyces cerevisiae'' [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=6320147 '''Mbp1'''] and defined its APSES (KilA-N) domain. Let us now search for an orthologue of this sequence in ''[[Species list|Your Species]]''. More precisely, you should identify prtoteins that fulfill the '''Reciprocal Best Match''' criterion.
| + | ==(1) Preparations== |
| − | | |
| − | First, we need to '''define the sequence''' you will use to find Mbp1 homologues. Since Mbp1 contains the very widely distributed Ankyrin motifs, a BLAST search with full length sequences will pick up a large number of Ankyrin-repeat containing proteins that are otherwise unrelated to our query. We will instead search for homologues using only the APSES domain as a query. However, the Pfam definition of the APSES domain (or KilA-N family, as it is now called) does not cover the entire length of the domain that has been crystallized. Therefore, we will use the sequence of the crystallized protein instead of the Pfam alignment. One of the results of our analysis will be '''whether APSES domains in fungi all have the same length as the Mbp1 domain, or whether some are indeed much shorter, as sugested by the Pfam alignment.''' To remind you, here is the full sequence of the [http://www.pdb.org/pdb/explore/derivedData.do?structureId=1MB1 1MB1 structure] (Note that the C-terminal His<sub>6</sub> tag that has been added for purification is not part of the Mbp1 protein sequence.) ...
| |
| − | | |
| − | | |
| − | >PDB:1MB1
| |
| − | MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEVLKETHEKVQGGFGKYQGTWVPL
| |
| − | NIAKQLAEKFSVYDQLKPLFDFTQTDGSASPPPAPKHHHASKVDHHHHHH
| |
| − | | |
| − | | |
| − | ... and, for comparison, this is the corresponding alignment with the Pfam KilA-N model obtained from a '''[http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi RPS-BLAST]''' search of the above sequence against the '''[http://www.ncbi.nlm.nih.gov/cdd/ CDD database]''':
| |
| − | | |
| − | | |
| − | <span style="color:#700777;"> 10 20 30 40 50 60 70 80</span>
| |
| − | <span style="color:#700777;"> ....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|</span>
| |
| − | <b>1MB1</b> <span style="color:#229922;"> 19 </span><span style="color:#2233cc;">IHSTGS</span><span style="color:#ff4466;">I</span><span style="color:#2233cc;">MK</span><span style="color:#ff4466;">R</span><span style="color:#2233cc;">K</span><span style="color:#ff4466;">KD</span><span style="color:#2233cc;">DWV</span><span style="color:#ff4466;">NAT</span><span style="color:#2233cc;">HIL</span><span style="color:#ff4466;">KAA</span><span style="color:#2233cc;">NFA</span><span style="color:#ff4466;">K</span><span style="color:#888888;">a</span><span style="color:#2233cc;">KRTRI</span><span style="color:#ff4466;">L</span><span style="color:#2233cc;">EK</span><span style="color:#ff4466;">E</span><span style="color:#2233cc;">VL</span><span style="color:#ff4466;">KE</span><span style="color:#2233cc;">TH</span><span style="color:#ff4466;">E</span><span style="color:#2233cc;">KVQ</span><span style="color:#888888;">----------------</span><span style="color:#ff4466;">G</span><span style="color:#2233cc;">GF</span><span style="color:#ff4466;">G</span><span style="color:#2233cc;">KY</span><span style="color:#ff4466;">QGT</span><span style="color:#2233cc;">W</span><span style="color:#ff4466;">V</span><span style="color:#2233cc;">PLNI</span> <span style="color:#229922;">82</span>
| |
| − |
| |
| − | Cdd:pfam04383 <span style="color:#229922;"> 3 </span><span style="color:#2233cc;">YNDFEI</span><span style="color:#ff4466;">I</span><span style="color:#2233cc;">IR</span><span style="color:#ff4466;">R</span><span style="color:#2233cc;">D</span><span style="color:#ff4466;">KD</span><span style="color:#2233cc;">GYI</span><span style="color:#ff4466;">NAT</span><span style="color:#2233cc;">KLC</span><span style="color:#ff4466;">KAA</span><span style="color:#2233cc;">GAT</span><span style="color:#ff4466;">K</span><span style="color:#888888;">-</span><span style="color:#2233cc;">RFRNW</span><span style="color:#ff4466;">L</span><span style="color:#2233cc;">RL</span><span style="color:#ff4466;">E</span><span style="color:#2233cc;">ST</span><span style="color:#ff4466;">KE</span><span style="color:#2233cc;">LI</span><span style="color:#ff4466;">E</span><span style="color:#2233cc;">ELS</span><span style="color:#888888;">kennidvliievenkk</span><span style="color:#ff4466;">G</span><span style="color:#2233cc;">KN</span><span style="color:#ff4466;">G</span><span style="color:#2233cc;">RL</span><span style="color:#ff4466;">QGT</span><span style="color:#2233cc;">Y</span><span style="color:#ff4466;">V</span><span style="color:#2233cc;">HPDL</span> <span style="color:#229922;">81</span>
| |
| − |
| |
| − |
| |
| − | <span style="color:#700777;"> 90</span>
| |
| − | <span style="color:#700777;"> ....*....|....*</span>
| |
| − | <b>1MB1</b> <span style="color:#229922;"> 83 </span><span style="color:#ff4466;">A</span><span style="color:#2233cc;">KQL</span><span style="color:#ff4466;">A</span><span style="color:#888888;">----</span><span style="color:#2233cc;">EK</span><span style="color:#ff4466;">F</span><span style="color:#2233cc;">SVY</span> <span style="color:#229922;">93</span>
| |
| − |
| |
| − | Cdd:pfam04383 <span style="color:#229922;"> 82 </span><span style="color:#ff4466;">A</span><span style="color:#2233cc;">LAI</span><span style="color:#ff4466;">A</span><span style="color:#888888;">swis</span><span style="color:#2233cc;">PE</span><span style="color:#ff4466;">F</span><span style="color:#2233cc;">ALK</span> <span style="color:#229922;">96</span>
| |
| − | | |
| − | | |
| − | As you can see, the Pfam alignment is 18 amino acids shorter at the N-terminus and 31 amino acids shorter at the C-terminus.
| |
| − | | |
| − | | |
| − | ;Find APSES domain proteins in your species:
| |
| − | | |
| − | <div style="padding: 5px; background: #EEEEEE;"> | |
| − | #Access the [[Species list|species list]] and identify the species that has been assigned to you.
| |
| − | #Navigate to the [http://www.ncbi.nlm.nih.gov '''NCBI's main page'''].
| |
| − | #In the left-hand menu of links, follow the link to [http://www.ncbi.nlm.nih.gov/guide/genomes-maps/ '''Genomes & Maps''']. | |
| − | #Under the '''Databases''' tab, follow the link to [http://www.ncbi.nlm.nih.gov/genome '''Genome'''].
| |
| − | #In the '''Genome tools''' section of that page, follow the link to [http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?species=euk '''Genomic groups BLAST'''].
| |
| − | #Click on link to the '''eukaryotic''' genomes tree, then on the link for the '''text table'''. This produces a BLAST interface to a list of species for which whole-genome sequences have been sequenced, annotated and entered into the various databases.
| |
| − | #Paste the FASTA sequence of the structurally defined Mbp1 APSES domain (e.g. from [http://www.pdb.org/pdb/explore/derivedData.do?structureId=1MB1 1MB1]) into the search field (excluding the His-tag, of course), set the parameters correctly for a '''Protein''' search against '''Protein''' sequences using '''blastp'''. Then find your [[Species list|assigned species]] in the table and check the box next to its name. Remember to record the parameters for your search. I expect you to understand which parameters would be needed in order to make this search reproducible. Run the search.
| |
| − | #On the next screen, check the box next to '''Format for: PSI-BLAST'''. Then click on '''View report''' to show the results of the first PSI-BLAST iteration.
| |
| − | #Run subsequent iterations of PSI-BLAST simply by clicking on '''Go''' after checking the sequences that have been included.
| |
| − | #Iterate the PSI-BLAST search until convergence (i.e. until no more '''new''' sequences are added); make sure to include only sequences for which the E-value is small (smaller than about 10e-03 should be safe). Sequences with borderline E-values that improve significantly in an iteration are probably homologues. Sequences with borderline E-values that do not improve much, or for which the E-value increases are probably not homologues. If this step does not work for you or the results are not what you expect, please contact your TA right away.
| |
| − | | |
| − | *Note: Please spend a little time on each page to understand its contents. <small>Ask, if the page contains resources or features you don't understand. Think about what you are doing. If you simply click on the links I provide, you will miss the opportunity to understand how the resources fit into the workflow you are working on, and to be able to execute similar processes yourself. Questions on page contents can potentially appear on quizzes and exam.</small>
| |
| − | </div>
| |
| − | | |
| − | | |
| − | Familiarize yourself with the '''output form''' you obtain, this is by far the most frequently used bioinformatics result page. You may want to refer to the [http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/new_view.html NCBI explanation].
| |
| − | | |
| − | Here is a list of things to look for, all of which I expect you to know and understand. (However you do not need to comment on these points in your submission.)
| |
| − | | |
| − | ;On the alignment image:
| |
| − | *What do the different colored bars mean?
| |
| − | *What is the information you get when you "mouse-over" a colored bar on the alignment image.
| |
| − | *What happens when you click on one of the bars?
| |
| − | | |
| − | ;In the description list:
| |
| − | *Where does the link next to an identifier take you?
| |
| − | *Where does the link in the "score" column take you?
| |
| − | *What does the icon at the end of each row mean? What other icons could appear there? <!-- cf. [http://www.ncbi.nlm.nih.gov/staff/tao/URLAPI/new_view.html] -->
| |
| − | | |
| − | ;In the alignment section:
| |
| − | *What do the alignment metrics mean:
| |
| − | **Score?
| |
| − | **Expect (E-value)?
| |
| − | **Identities?
| |
| − | **Positives?
| |
| − | **Gaps?
| |
| − | *What is the alignment length?
| |
| − | *Which sequence is labeled '''Query''' and which one is labelled '''Sbjct'''?
| |
| − | | |
| − | | |
| − | ;Next
| |
| − | :retrieve the sequences that have E-values low enough to make you conclude they contain APSES domain homologues.
| |
| − | | |
| − | <div style="padding: 5px; background: #EEEEEE;">
| |
| − | | |
| − | #Review the sequences you have found: they should all be significantly similar to the query profile. In some of the assigned species you will find one hit for each distinct sequence in the genome, in others, you will find several versions of essentially the same gene (e.g. refseq and other accession numbers).
| |
| − | #Explore the relationship between the hits by clicking on '''select all sequences''', then choosing '''Distance tree of results''' at the top or bottom of your search results to visualize a tree representation of similarity. Highly similar sequences will be collapsed into the same node in the distance tree; you can expand those nodes to list all the node's members.
| |
| − | #Identify '''one''' representative for each distinct protein you have found. If possible, use proteins with refseq identifiers. Avoid duplicates or nearly identical variants. If there are length differences, use the longer version (shorter versions may contain only partial sequences). Click on the checkbox next to each protein you have identified.
| |
| − | #Click on '''get selected sequences''' at the top or bottom of the page. Note and record the GIs for your sequences that are listed in the ''Search details'' box, you can use them to easily reproduce your results by pasting them into any Entrez search. Also note the URL that this has produced (in your browser's URL bar). As you see, you can retrieve a list of sequences from NCBI simply by adding a list of comma-separated GI numbers to the [http://www.ncbi.nlm.nih.gov/protein/ URL of the protein database].
| |
| − | #Click on '''Display settings''' and choose '''FASTA (text)'''.
| |
| − | | |
| − | <small>If you want, for comparison, you can run a multiple alignment with an NCBI-developed MSA tool: '''COBALT'''. On the sequence list page, in the right-hand column, in the section '''Analyze these sequences''', click on '''Align sequences with COBALT'''. It is a convenient way to get a quick first look at an alignment of NCBI retrieved sequences.</small>
| |
| | </div> | | </div> |
| − | | + | |
| − | You now have a collection of APSES domain-containing homologues in your organism. There are two more tasks we need to address before we can compute alignments and analyze them. (A) we need to rename our sequences, and (B) we need to define the boundaries of their APSES domains.
| + | |
| − | | |
| − | | |
| | | | |
| | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> |
| − | | + | ===(1.1) Preparing Input Files=== |
| − | ===(1.2) Renaming Sequences=== | |
| | </div> | | </div> |
| | + | <br> |
| | | | |
| − | A phylogenetic tree or multiple alignment is not really informative if it that displays GI numbers or other abstract identifiers as labels of rows or nodes. The relationship between species is fundamental to the variation we observe and we need to make this relationship explicit.
| + | =====Introduction: Task===== |
| | + | For this assignment, we start from the multiple sequence alignments we have constructed previously. We will edit the alignment to make it suitable for phylogenetic analysis. We will construct a phylogenetic tree and we will analyse the tree. |
| | | | |
| − | Imagine that the rows in an MSA were completely unlabeled, or the nodes in the tree would be just circles: we would have a very hard time relating the computed relationships back to the biology they represent. Abstract identifiers like <tt>NP_010227</tt> are not much better.
| + | The phylogenetic tree we will construct will represent all APSES domains of the species we have analyzed. In order to '''interpret''' such a tree it is crucial to have some sense of what these domains are, i.e. to cluster them according to their orthologues. Only then can we analyse the tree by asking which subclades mirror the accepted phylogeny of fungi and which ones differ. In the third assignment, we have assigned orthology from reciprocal best match analysis. Based on this information, I have revised the gene names in the [[APSES_domains_MUSCLE_revised|'''MUSCLE alignment of all APSES domains''']]. When we calculate a phylogenetic tree with these sequences, we should expect orthologues to cluster into the same subclade. Of course, not all fungi have the same number of APSES domain homologues, but from the data we have compiled it should be possible to define their evolutionary history with reference to the other species. |
| | | | |
| − | Typically, the information that programs use to label sequences is taken from the FASTA header. This provides us with an easy way to make sure they display the information we need and that we can interpret. Typically such programs will use the first few (often ten) characters they find. We will therefore design short strings strings that identify potential gene family relationships as well as species.
| + | =====Introduction: Principle===== |
| | + | In order to use molecular sequences for the construction of phylogenetic trees, you have to build a multiple alignment first, then edit it. This is important: all rows of sequences have to contain the exact same number of characters and to hold '''aligned characters in corresponding positions'''. Phylogeny programs are not meant to revise an alignment but to analyse evolutionary relationships, given the alignment. Their inferences are made on a column-wise basis and if your columns contain data from unrelated positions, the inferences are going to be questionable. |
| | | | |
| | + | The result of the tree construction is a decision about the most likely evolutionary relationships. Fundamentally, tree-construction programs decide which sequences had common ancestors. |
| | | | |
| − | ;Species codes
| + | '''Distance based''' phylogeny programs start by using sequence comparisons to estimate evolutionary distances: |
| | + | * they apply a model of evolution such as a mutation data matrix, to calculate a score for each '''pair''' of sequences, |
| | + | * this score is stored in a "distance matrix" ... |
| | + | * ... and used to estimate a tree that goups sequences with close relationships together. (e.g. by using an NJ, Neigbor Joining, algorithm). |
| | + | They are fast, can work on large numbers of sequences, but are less accurate if genes evolve at different rates. |
| | | | |
| − | The scientific name of a species is formed according to Linnaean [http://en.wikipedia.org/wiki/Binomial_nomenclature binomial nomenclature] and Swissprot has for a long time condensed species names into mnemonic five-character codes, taking the first three from the [http://en.wikipedia.org/wiki/Genus genus name] and the last two from the [http://en.wikipedia.org/wiki/Specific_name specific name]. For example ''Saccharomyces cerevisiae'' is abbreviated as <tt>SACCE</tt> and ''Lachancea thermotolerans'' is <tt>LACTH</tt>. For the most part, this creates unique strings that are good mnemonic labels for the species. I have added these "codes" to the [[Species list]].
| + | '''Parsimony based''' phylogeny programs build a tree that minimizes the number of mutation events that are required to get from a common ancestral sequence to all observed sequences. They take all columns into account, not just a single number per sequence pair, as the Distance Methods do. For closely related sequences they work very well, but they construct inaccurate trees when they can't make good estimates for the required number of sequence changes. |
| | | | |
| | + | '''ML''', or '''Maximum Lieklihood''' methods attempt to find the tree for which the observed sequences would be the most likely under a particular evolutionary model. They are based on a rigorous statistical framework and yield the most robust results. But they are also VERY compute intensive and a tree of the size that we are building in this assignment is already almost beyond the resources of common workstations (runs about a day on my computer). However, one may split a large problem into smaller, obvious subtrees (e.g. analysing orthologues as a group, only including a few paralogues for comparison) and then merge the smaller trees; this way even very large problems can become tractable. They also suffer less from "long-branch attraction" - the phenomenon that weakly similar sequences can be grouped inappropriately close together in a tree due to spurious shared differences. |
| | | | |
| − | ;Gene families
| + | Clearly, in order for tree-estimation to work, one must not include fragments of sequence which have evolved under a different evolutionary model as all others, e.g. after domain fusion, or after accommodating large stretches of indels. Thus it is appropriate to edit the sequences and pare them down to a ''most characteristic subset'' of amino acids. The goal is not to be as comprehensive as possible, but to input those columns of aligned residues that will best represent the ''true'' phylogenetic relationships between the sequences. |
| − | Most yeast genes have traditional names, like mbp1 or sok2. These names are convenient family labels since ''saccharomyces cerevisiae'' is one of the best studied [http://en.wikipedia.org/wiki/Model_organism model organisms]. Therefore, once we identify a protein family that includes a yeast gene, we can easily access expert knowledge in textbooks or manuscripts. Of course, such labels are arbitrary - whether we call a gene '''Mbp1''' or '''WXYZ''' makes no difference - as long as all genes that we presume to be family members carry the same label. For higher eukaryotes, I would probably choose human gene names as a reference point, for bacteria I would choose ''E. coli''.
| |
| | | | |
| − | To define which gene belongs into which family, we can align all newly found genes with all yeast APSES domain homologues, to find out which ones they are most similar to. This creates common family labels. We can use these as provisional family names for the encoded proteins, even though we may want to revise them once we have mapped out explicit phylogenetic trees.
| + | =====Introduction: Problems===== |
| | + | Gaps are a real problem here, as usual. Strictly speaking, the similarity score of an '''alignment''' program as well as the distance score of a '''phylogeny''' program are not calculated for an ordered ''sequence'', but for a ''sum of independent values'', one for each aligned columns of characters. The order of the columns does not change the score. Hoever in an optimal sequence alignment with gaps, this is no longer strictly true since a one-character gap creation has a different penalty score than a one-character gap extension! Most '''alignment''' programs use a model with a constant gap insertion penalty and a linear gap extension penalty. This is not rigourously justified from biology, but parametrized (or you could say "tweaked") to correspond to our observations. However, most '''phylogeny''' programs, (such as the programs in PHYLIP) do not work in this way. PHYLIP strictly operates on columns of characters and treats a gap character just like a residue with the one letter code "-". Thus gap insertion- and extension- characters get the samescore. For short indels, this '''underestimates''' the distance between pairs of sequences, since any evolutionary model should reflect the fact that gaps are much less likely than point mutations. If the gap is very long though, all events are counted individually as many single substitutions (rather than one lengthy one) and this '''overestimates''' the distance. And it gets worse: long stretches of gaps can make sequences appear similar in a way that is not justified, just because they are identical in the "-" character. It is therefore common and acceptable to edit gaps in the alignment to one or two character, or to remove them. |
| | | | |
| | + | =====Introduction: Practice===== |
| | + | In practice, follow the fundamental principle that '''all characters in a column should be related by homology'''. This implies the following rules of thumb: |
| | | | |
| − | ;Identifying APSES domains (general procedure).
| + | :*Remove all stretches of residues in which the ''alignment'' appears ambiguous (not just highly variable, but ambiguous regarding the aligned positions). |
| − | In order to identify the APSES domain boundaries, you can simply run a multiple sequence alignment of the structurally defined APSES domain sequence (e.g. taken from PDB-ID 1MB1) against all sequences you have found. The boundaries of the aligned APSES domain then define the domain boundaries in the aligned proteins.
| + | :*Remove all frayed N- and C- termini, especially regions in which not all sequences that are being compared appear homologous and that may stem from unrelated domains. |
| | + | :*Remove all but approximately one column from gapped regions, and all residues N- and C- terminal of the gap in which the alignment appears questionable. ( I would keep one gapped column as a placeholder for a rare and very distinct evolutionary event, rather than simply deleting them all, some researchers remove all gaps). |
| | + | :*Also, consider that neither residues that are completely different between all species, nor residues that are completely conserved are informative for relationship distances. |
| | + | :*If your sequences are too long, you may run out of memory. 60-80 aligned residues should be plenty and if the sequences fit on a single line you will save yourself potential trouble with block-wise vs. interleaved input. |
| | | | |
| | + | :<small>(A '''very''' useful trick with Microsoft Word is that you can select blocks of text and entire columns in the document with your mouse: hold the "ALT" key depressed while you click and drag your mouse to select. This will greatly facilitate the preparation of sequences. You can treat that selection as any other selected text: color or highlight characters, or delete them. Importantly, you can also cut and paste entire columns! Of course, this will only work as expected if you use a fixed-width font such as Courier or "Courier New". )</small> |
| | | | |
| − | ;Identifiying family relationships (in the same run) | + | The preparation of the input file of aligned residues, used by the PHYLIP package is straightforward in principle; just carefully follow the instructions in PHYLIP's well written documentation. If you plan to use an outgroup for your tree, it is a good idea to move that to the first line of your alignment, since this is where PHYLIP will look for it by default. |
| − | However, for efficiency, we can also determine '''family relationships''' in the same alignment that we use to define domain boundaries, if we simply include '''all''' yeast APSES domains in the MSA. Then we can judge similarity simply from examining the guide tree of the alignment and label the families accordingly. This has the added advantage that the domain boundaries are more securely defined, since we include more sequence information into the alignment.
| |
| | | | |
| − | ;Proceed as follows. | + | Some notes on how to avoid common editing troubles. Copy the sequences from the pages linked from the ''Resources'' section below. Paste them into a document, using the Word "Edit → Paste special → Unformatted text". Set the page-setup to "landscape", the font-size to something small, then you can put every sequence into one line. Take special note that your files must not include tab characters! (Tabs are counted as one single character by the phylogeny programs.) You can use Word to globally replace all tabs (specified as "^t") with a blank, to make sure. Spaces count, so display your alignment in a fixed-width font, such as Courier (or "Courier New"), not a proportional-width font such as Times, Arial, or Helvetica, and ensure all columns in your alignments align as they should. As always, make sure you save your input files as "Text Only". |
| | | | |
| − | <div style="padding: 5px; background: #EEEEEE;"> | + | <small> |
| − | #Open the [http://www.ebi.ac.uk/Tools/muscle/ Muscle MSA input page] at the EBI.
| + | :A note if you are working on a '''Mac''' and saving input on disk, to run with a locally installe PHYLIP version: here MS Word will play one of its usual [http://en.wikipedia.org/wiki/Shenanigan shenanigans] on you since it writes text files with the old-style OS 9 Carriage Return characters <code>(\r; ASCII 13; hex 0D; CR)</code>. Just by looking at the file, this is quite invisible but such "Carriage returns" are not going to be recognized by PHYLIP and most other UNIX based programs. It may not make a difference when you paste your sequences to a Web server; but if you compute things locally it will appear to the program as though all the input would be passed in one single, very long line). And this can (and did) lead to head-banging rounds of frustration. You need to replace them with '''Linefeed''' resp. '''Newline''' characters <code>(\n; ASCII 10; hex 0A; LF)</code> and you can't even do that within Word(!). Open a UNIX terminal window and navigate to the directory where your files reside. Then type: |
| − | #Access the [[APSES domains (yeast)|Yeast APSES domain collection]] I have prepared and copy the FASTA sequences. Paste them into the sequence field of the MUSCLE program input form.
| |
| − | #Copy the FASTA sequenced of the full length APSES domain protein sequence collection from your PSI-BLAST search (above) and paste them into the MUSCLE input form as well.
| |
| − | #Set the following parameters:
| |
| | | | |
| − | OUTPUT FORMAT: CLUSTALW2
| + | :'''tr "\r" "\n" < infile > outfile''' |
| − | OUTPUT TREE: from second iteration
| |
| − | OUTPUT ORDER: aligned
| |
| | | | |
| − | #Click on Submit.
| + | :... where outfile is different from infile (careful: if a file by the name of outfile already exists, '''tr''' will cheerfully overwrite it.) Alternatively you could type the following perl one-line program : |
| − | </div>
| |
| | | | |
| | + | :'''perl -e 'while(<>){tr/\r/\n/;print}' < infile > outfile''' |
| | + | </small> |
| | | | |
| − | The output should show the MSA. The overlap of the yeast APSES domains with your sequences defines the domain boundaries. Moreover, a tree has been calculated and you can view the tree to identify family relationships.
| |
| | | | |
| − | ;Visualize the alignment tree and decide on names
| + | In your assignment submission, clearly highlight or otherwise color the columns that you have selected, annotate why you have selected them and paste your resulting input file as well. Here is an example of what this might look like: |
| | | | |
| − | <div style="padding: 5px; background: #EEEEEE;">
| |
| − | Click on the link to the Guide tree. This is the so-called Newick tree format and there are a large number of online tree viewers to visualize such trees. The MUSCLE form will display one tree for you,
| |
| | | | |
| − | <small>You could also navigate (for example) to the [http://www.proweb.org/treeviewer/ proWeb Tree viewer] and paste the tree data into the '''User-supplied Newick Tree''' input field. Choose any graphics format your browser can handle (JPEG is a pretty safe bet) and click on '''View tree'''.</small>
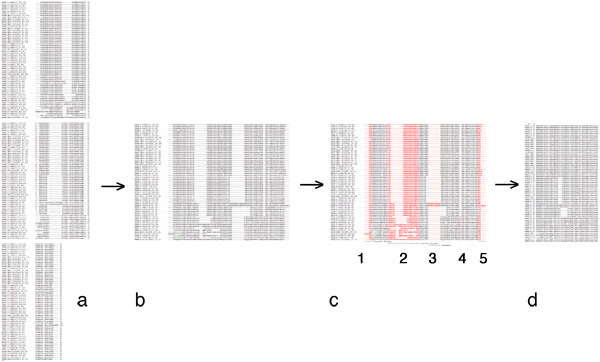
| + | [[Image:EditingGuide.jpg|frame|none|(Possible) steps in editing a multiple sequence alignment towards a PHYLIP input file. '''a''': raw alignment (CLUSTAL format); '''b''': sequences assembled into single lines; '''c''': columns to be deleted highlighted in red - 1, 3 and 4: large gaps; 2: uncertain alignment and 5: frayed C-terminus: both would put non-homologous characters into the same column; '''d''': input data for PHYLIP: names for sequences must not be longer than 10 characters, the first line must contain the number of sequences and the sequence length. PHYLIP is very picky about incorrectly formatted input, read the [http://evolution.genetics.washington.edu/phylip/doc/sequence.html PHYLIP sequence format guide].]] |
| | | | |
| | + | =====Introduction: Web Service and data===== |
| | | | |
| − | #Interpret the tree to decide on the protein family names for your sequences:
| + | You have two choices for completing the assignment: either to use one of the [http://evolution.gs.washington.edu/phylip/phylipweb.html PHYLIP on-line servers] that generously provide public computing resources, or to download and install the [http://evolution.genetics.washington.edu/phylip.html PHYLIP program package] on your own computer at home. If you choose the former, one of your options is the [http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html '''PHYLIP''' service at the '''Institut Pasteur'''] in France. |
| − | ##If a yeast protein is grouped with exactly one of your proteins, your protein gets the same name.
| |
| − | ##If a yeast protein is grouped with more than one of your proteins, replace the number in the yeast protein with a, b, c ..., from most similar to least similar for your protein. For example: if one Aspergillus fumigatus protein is most similar to yeast Mbp1, you will give it the name MBP1_ASPFU. If two proteins are both most similar to yeast Sok2, you will name them SOKA_ASPFU and SOKB_ASPFU. Try to get it approximately right but remember that this is a process of estimation - we are not accurately measuring distances (yet).
| |
| | | | |
| − | That done, edit your FASTA headers and save your APSES domain sequence set. We will need them for the next assignment.
| + | <small>I have tried the Pasteur service many times, and it works - however not always entirely without problems. Uninformative errors may occur when your input is too large for the system's memory (like: "sequences not aligned" ... "out of memories" and such) and once, after submitting a number of jobs, the system locked me out to wait until results would be received by e-mail (which then hasn't happened). Regrettably, this is not documented. However the integration of their services in a logical sequence of steps is very convenient and some of their services use algorithms that improve on PHYLIP. If you rather decide to install PHYLIP, good for you. That is easy to do, well documented, there are much less limitations on memory - but if you don't read and understand the instructions carefully, you may be in for a spell of frustration.</small> |
| | | | |
| − | </div>
| + | Either way, I have posted typical input files and result files on the [[Assignment_5_fallback_data|fallback data page]], to allow you to bail out in case technical problems become overwhelming. If you use the data posted here instead of your own, you '''must''' document that fact and explain what you have tried, and why that has failed. The posted data is a fallback, not a shortcut. |
| | | | |
| | + | For this assignment, we will use a simple distance based tree construction method, specifically the UPGMA variant of the neighbor joining algorithm. This represents a reasonable compromise between accuracy and speed, especially when applied to moderately dissimilar sequences. In general, distance methods include '''two''' steps: (1) calculate a pairwise-distance matrix between sequences, (2) construct a tree, based on the matrix. Thus all the information in the alignment bewtween two pairs of sequences is collapsed into a single number: their pairwise distance. Alternative approaches, parsimony as well as ML based algorithms, take individual columns into account. |
| | | | |
| | + | <br> |
| | + | <div style="padding: 5px; background: #DDDDEE;"> |
| | + | Prepare an input file that is representative of the APSES domains. |
| | | | |
| − | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;">
| + | *Access the [[APSES_domains_MUSCLE_revised|revised MSA for all APSES domains]], linked here (and from the resources section at the bottom of the page). Prepare a PHYLIP formatted input file from this MSA, restricting the number of sequence characters to no more than 70. Read the [http://evolution.genetics.washington.edu/phylip/doc/main.html#inputfiles PHYLIP format documentation] and follow the considerations dicussed above. ([[Assignment_5_fallback_data|See the fallback data in case you get stuck]], but you '''must''' prepare (and document) an input file according to the instructions, even if you end up using the fallback data for whatever reason.) Do not forget to document how you have prepared your input file: define where your source-sequences came from, define which columns you have deleted by highlighting the deleted residues in one sequence, and include your input file in the assignment. |
| − | | |
| − | ==(2) Align and Annotate==
| |
| | </div> | | </div> |
| | | | |
| | <br> | | <br> |
| − | | + | <br> |
| | | | |
| | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> |
| − | ===(2.1) Review of domain annotations===
| |
| − | </div>
| |
| − |
| |
| − | APSES domains are relatively easy to identify and annotate but we have had problems with the ankyrin domains in Mbp1 homologues. Both CDD as well as SMART have identified such domains, but while the domain model was based on the same Pfam profile for both, and both annotated approximately the same regions, the details of the alignments and the extent of the predicted region was different.
| |
| − |
| |
| − | [http://www.yeastgenome.org/cgi-bin/locus.fpl?locus=mbp1 Mbp1] forms heterodimeric complexes with a homologue, [http://www.yeastgenome.org/cgi-bin/locus.fpl?locus=swi6 Swi6]. Swi6 does not have an APSES domain, thus it does not bind DNA. But it is similar to Mbp1 in the region spanning the ankyrin domains and in [http://www.ncbi.nlm.nih.gov/pubmed/100489281999 Foord et al. published] its crystal structure ([http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1SW6 1SW6]). This structure is a good model for Ankyrin repeats in Mbp1. For details, please refer to the consolidated [[Mbp1 annotation|Mbp1 annotation page]] I have prepared.
| |
| − |
| |
| − | In what follows, we will use the program JALVIEW - a Java based multiple sequence alignment editor to load and align sequences and to consider structural similarity between yeast Mbp1 and its closest homologue in your organism.
| |
| − |
| |
| − | In this part of the assignment,
| |
| − |
| |
| − | #You will load sequences that are most similar to Mbp1 into an MSA editor;
| |
| − | #You will add sequences of ankyrin domain models;
| |
| − | #You will perform a multiple sequence alignment;
| |
| − | #You will try to improve the alignment manually;
| |
| − | <!-- Finally you will consider if the Mbp1 APSES domains could extend beyond the section of homology with Swi6 -->
| |
| − |
| |
| | | | |
| − | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;">
| + | ===(1.2) Calculating a Tree=== |
| − | ===(2.2) Jalview, loading sequences=== | |
| | </div> | | </div> |
| | | | |
| − | Geoff Barton's lab in Dundee has developed an integrated MSA editor and sequence annotation workbench with a number of very useful functions. It is written in Java and should run on Mac, Linux and Windows platforms without modifications. We will use this tool for this assignment and explore its features as we go along.
| + | <br> |
| − | | + | <br> |
| − | <div style="padding: 5px; background: #EEEEEE;"> | + | <div style="padding: 5px; background: #DDDDEE;"> |
| − | #Navigate to the [http://www.jalview.org/ Jalview homepage] click on '''Download''', install Jalview on your computer and start it. A number of windows that showcase the program's abilities will load, you can close these.
| |
| − | #Prepare homologous Mbp1 sequences for alignment:
| |
| − | ##Find the sequence in your assigned species that fulfills the Reciprocal Best Match crierion with yeast Mbp1.
| |
| − | ##Open the [[Mbp1 RBM reference sequences]] page.
| |
| − | ##Copy the FASTA sequences of the reference proteins, return to Jalview and select File → Input Alignment → from Textbox and paste the sequences into the textbox.
| |
| − | ##Also paste a FASTA sequence of your species' Mbp1 protein into the window.
| |
| − | ##Finally copy the sequences for ankyrin domain models (below) and paste them into the Jalview textbox as well. Paste two separate copies of the CD00204 consensus sequence and one copy of 1SW6.
| |
| − | ##When all the sequences are present, click on New Window. Jalview gives you all the sequences, but of course this is not yet an alignment.
| |
| − | | |
| − | </div>
| |
| − | | |
| − | ;Ankyrin domain models
| |
| − | >CD00204 ankyrin repeat consensus sequence from CDD
| |
| − | NARDEDGRTPLHLAASNGHLEVVKLLLENGADVNAKDNDGRTPLHLAAKNGHLEIVKLLL
| |
| − | EKGADVNARDKDGNTPLHLAARNGNLDVVKLLLKHGADVNARDKDGRTPLHLAAKNGHL
| |
| | | | |
| − | >1SW6 from PDB - unstructured loops replaced with xxxx
| + | *Using the '''protdist''' program of PHYLIP, calculate a distance matrix for the input file you have prepared. ([[Assignment_5_fallback_data|See the fallback data in case you get stuck]]) (1 mark) |
| − | GPIITFTHDLTSDFLSSPLKIMKALPSPVVNDNEQKMKLEAFLQRLLFxxxxSFDSLLQE
| |
| − | VNDAFPNTQLNLNIPVDEHGNTPLHWLTSIANLELVKHLVKHGSNRLYGDNMGESCLVKA
| |
| − | VKSVNNYDSGTFEALLDYLYPCLILEDSMNRTILHHIIITSGMTGCSAAAKYYLDILMGW
| |
| − | IVKKQNRPIQSGxxxxDSILENLDLKWIIANMLNAQDSNGDTCLNIAARLGNISIVDALL
| |
| − | DYGADPFIANKSGLRPVDFGAG
| |
| | | | |
| | + | *If you use the PHYLIP Webserver, select the neighbor joining algorithm from the menu options ('''neighbor''' on the PHYLIP server) and click the button "run the selected program on outfile" ; on the next form, click the button to the "advanced neighbor form", choose the option "UPGMA" and click on the button "run neighbor". When the program is done, select the option '''drawgram''' and click '''Run the selected program on outtree'''. Choose a '''cladogram''' tree-style and a suitable output format (e.g. postscript). Paste the trees into your assignment. |
| | | | |
| | + | *If you use a locally installed version of PHYLIP use '''neighbor''' with the UPGMA method to construct a tree for the input file. Open the file '''outfile''' in a text-editor, copy and paste the trees into your assignment. |
| | | | |
| − | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | + | In both cases, the process is: <code>protdist</code> → <code>neighbor</code> → <code>drawgram</code> |
| | | | |
| − | ===(2.3) Computing alignments===
| |
| | </div> | | </div> |
| | | | |
| − | Sequence alignments can be calculated directly from Jalview.
| + | <br> |
| − | | + | <br> |
| − | <div style="padding: 5px; background: #EEEEEE;"> | |
| − | #In Jalview, select '''Web Service → Alignment → MAFFT Multiple Protein Sequence Alignment'''. The alignment is calculated in a few minutes and displayed in a new window.
| |
| − | #Choose '''Colour → Hydrophobicity''' and '''→ by Conservation'''. Then select '''Modify Conservation Threshold...''' and adjust the slider left or right to see which columns are highly conserved. You will notice that the Swi6 sequence that was supposed to align only to the ankyrin domains was in fact aligned to other parts of the sequence as well. This is one part of the MSA that we will have to correct manually and a common problem when aligning sequences of different lengths.
| |
| − | #Other alignment algorithms are available and you may wish to explore whether the alignments differ significantly.
| |
| − | </div> | |
| | | | |
| | + | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> |
| | | | |
| − | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;">
| + | ==(2) Analysis (3 marks)== |
| − | ===(2.4) Editing ankyrin domain alignments===
| |
| | </div> | | </div> |
| | | | |
| | + | I have constructed a cladogram for the species we are analysing, based on data published for 1551 fungal ribosomal sequences. Such reference tres from rRNA data are a standard method of phylogenetic analysis, supported by the assumption that rRNA sequences are monophyletic and have evolved under comparable selective pressure in all species. |
| | | | |
| − | A '''good''' MSA comprises only columns of residues that play similar roles in the proteins' mechanism and/or that evolve in a comparable structural context. Since it is a result of biological selection and conservation, it has relatively few indels and the indels it has are usually not placed into elements of secondary structure or into functional motifs. The contiguous features annotated for Mbp1 are expected to be left intact by a good alignment.
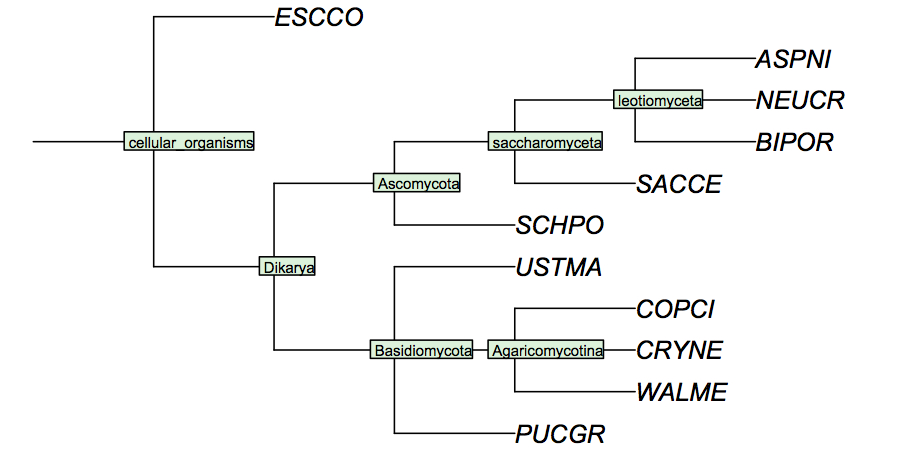
| + | [[Image:FungiCladogram.jpg|frame|none|Cladogram of fungi studied in the assignments. This cladogram is based on small subunit ribosomal rRNA sequences, and largely follows ''Tehler et al.'' (2003) ''Mycol Res.'' '''107''':901-916. Even though many details of fungal phylogeny remain unresolved, the branches shown here individually appear to have strong support. In a cladogram such as this, the branch lengths are not drawn to any scale of similarity. I have labeled all speciation events so you can refer to these labels in your assignment.]] |
| | | | |
| − | A '''poor''' MSA has many errors in its columns; these contain residues that actually have different functions or structural roles, even though they may look similar according to a (pairwise!) scoring matrix. A poor MSA also may have introduced indels in biologically irrelevant positions, to maximize spurious sequence similarities. Some of the features annotated for Mbp1 will be disrupted in a poor alignment and residues that are conserved may be placed into different columns.
| + | In order to study the evolutionary history of the entire gene family you can use the tree you have computed or access the [[APSES_domains_reference_tree|'''APSES domains reference tree''']] here. |
| | | | |
| − | Often errors or inconsistencies are easy to spot, and manually editing an MSA is not generally frowned upon, even though this is not a strictly objective procedure. The main goal of manual editing is to make an alignment biologically more plausible. Most comonly this means to mimize the number of rare evolutionary events that the alignment suggests and/or to emphasize conservation of known functional motifs. Here are some examples for what one might aim for in manually editing an alignment:
| + | This is a complicated tree, and it can look impenetrably confusing at first. Here are two principles that will help you make sense of the tree. |
| | | | |
| − | ;Reduce number of indels
| + | A: '''A gene that is present in an ancestral species, is inherited in all descendent species.''' The gene has to be observed in all OTUs, unless its has been lost (which is a rare event). This means, if a gene is present in two widely divergent species, but in none other of the descendants of the LCA, it is possible that there is some problem with the tree (long branch attraction maybe), or the sequence has been acquired through horizontal gene transfer. |
| − | From a Probcons alignment:
| |
| − | 0447_DEBHA ILKTE-K<span style="color: rgb(255, 0, 0);">-</span>T<span style="color: rgb(255, 0, 0);">---</span>K--SVVK ILKTE----KTK---SVVK
| |
| − | 9978_GIBZE MLGLN<span style="color: rgb(255, 0, 0);">-</span>PGLKEIT--HSIT MLGLNPGLKEIT---HSIT
| |
| − | 1513_CANAL ILKTE-K<span style="color: rgb(255, 0, 0);">-</span>I<span style="color: rgb(255, 0, 0);">---</span>K--NVVK ILKTE----KIK---NVVK
| |
| − | 6132_SCHPO ELDDI-I<span style="color: rgb(255, 0, 0);">-</span>ESGDY--ENVD ELDDI-IESGDY---ENVD
| |
| − | 1244_ASPFU ----N<span style="color: rgb(255, 0, 0);">-</span>PGLREIC--HSIT -> ----NPGLREIC---HSIT
| |
| − | 0925_USTMA LVKTC<span style="color: rgb(255, 0, 0);">-</span>PALDPHI--TKLK LVKTCPALDPHI---TKLK
| |
| − | 2599_ASPTE VLDAN<span style="color: rgb(255, 0, 0);">-</span>PGLREIS--HSIT VLDANPGLREIS---HSIT
| |
| − | 9773_DEBHA LLESTPKQYHQHI--KRIR LLESTPKQYHQHI--KRIR
| |
| − | 0918_CANAL LLESTPKEYQQYI--KRIR LLESTPKEYQQYI--KRIR
| |
| | | | |
| − | <small>Gaps marked in red were moved. The sequence similarity in the alignment does not change considerably, however the total number of indels in this excerpt is reduced to 13 from the original 22</small>
| + | B: '''Paralogous genes in an ancestral species should give rise to monophyletic subtrees for each of the genes, in all descendants'''; this means: if the LCA of a branch has e.g. three genes, we would expect three copies of the species cladogram below this branchpoint, one for each of these genes. Each of these subtrees should recapitulate the reference phylogenetic tree of the OTUs, up to the branchpoint of their LCA. |
| | | | |
| | + | With these two simple principles (you should draw them out on a piece of paper if they do not seem obvious to you), you can probably pry the [[APSES_domains_reference_tree|reference tree of all APSES domains]] apart quite nicely. A few colored pencils and a printout of the tree will help. |
| | | | |
| − | ;Move indels to more plausible position
| |
| − | From a CLUSTAL alignment:
| |
| − | 4966_CANGL MKHEKVQ------GGYGRFQ---GTW MKHEKV<span style="color: rgb(0, 170, 0);">Q</span>------GGYGRFQ---GTW
| |
| − | 1513_CANAL KIKNVVK------VGSMNLK---GVW KIKNVV<span style="color: rgb(0, 170, 0);">K</span>------VGSMNLK---GVW
| |
| − | 6132_SCHPO VDSKHP<span style="color: rgb(255, 0, 0);">-</span>----------<span style="color: rgb(255, 0, 0);">Q</span>ID---GVW -> VDSKHP<span style="color: rgb(0, 170, 0);">Q</span>-----------ID---GVW
| |
| − | 1244_ASPFU EICHSIT------GGALAAQ---GYW EICHSI<span style="color: rgb(0, 170, 0);">T</span>------GGALAAQ---GYW
| |
| | | | |
| − | <small>The two characters marked in red were swapped. This does not change the number of indels but places the "Q" into a a column in which it is more highly conserved (green). Progressive alignments are especially prone to this type of error.</small>
| + | |
| | + | |
| | | | |
| − | ;Conserve motifs
| |
| − | From a CLUSTAL alignment:
| |
| − | 6166_SCHPO --DKR<span style="color: rgb(255, 0, 0);">V</span>A---<span style="color: rgb(255, 0, 0);">G</span>LWVPP --DKR<span style="color: rgb(0, 255, 0);">V</span>A--<span style="color: rgb(0, 255, 0);">G</span>-LWVPP
| |
| − | XBP1_SACCE GGYIK<span style="color: rgb(255, 0, 0);">I</span>Q---<span style="color: rgb(255, 0, 0);">G</span>TWLPM GGYIK<span style="color: rgb(0, 255, 0);">I</span>Q--<span style="color: rgb(0, 255, 0);">G</span>-TWLPM
| |
| − | 6355_ASPTE --DE<span style="color: rgb(255, 0, 0);">I</span>A<span style="color: rgb(255, 0, 0);">G</span>---NVWISP -> ---DE<span style="color: rgb(0, 255, 0);">I</span>A--<span style="color: rgb(0, 255, 0);">G</span>NVWISP
| |
| − | 5262_KLULA GGYIK<span style="color: rgb(255, 0, 0);">I</span>Q---<span style="color: rgb(255, 0, 0);">G</span>TWLPY GGYIK<span style="color: rgb(0, 255, 0);">I</span>Q--<span style="color: rgb(0, 255, 0);">G</span>-TWLPY
| |
| | | | |
| − | <small>The first of the two residues marked in red is a conserved, solvent exposed hydrophobic residue that may mediate domain interactions. The second residue is the conserved glycine in a beta turn that cannot be mutated without structural disruption. Changing the position of a gap and insertion in one sequence improves the conservation of both motifs.</small> | + | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> |
| − | | |
| − | | |
| − | The Ankyrin domains are quite highly diverged, the boundaries not well defined and not even CDD, SMART and SAS agree on the precise annotations. We expect there to be alignment errors in this region. Nevertheless we would hope that a good alignment would recognize homology in that region and that ideally the required <i>indels</i> would be placed between the secondary structure elements, not in their middle. But judging from the sequence alignment alone, we cannot judge where the secondary structure elements ought to be. You should therefore add the following "sequence" to the alignment; it contains exactly as many characters as the Swi6 sequence above and annotates the secondary structure elements. I have derived it from the 1SW6 structure
| |
| − | | |
| − | >SecStruc 1SW6 E: strand t: turn H: helix _: irregular
| |
| − | _EEE__tt___ttt______EE_____t___HHHHHHHHHHHHHHHH_xxxx_HHHHHHH
| |
| − | HHHH_t_____t_____t____HHHHHHH__tHHHHHHHHH____t___tt____HHHHH
| |
| − | HH__HHHH___HHHHHHHHHHHHHEE_t____HHHHHHHHH__t__HHHHHHHHHHHHHH
| |
| − | HHHHHH__EEE_xxxx_HHHHHt_HHHHHHH______t____HHHHHHHH__HHHHHHHH | |
| − | H____t____t____HHHH___
| |
| − | | |
| − | | |
| − | To proceed:
| |
| − | #You should manually align the Swi6 sequence with yeast Mbp1 | |
| − | #You should bring the Secondary structure annotation into its correct alignment with Swi6
| |
| − | #You should bring both CDD ankyrin profiles into the correct alignment with yeast Mbp1
| |
| − | | |
| − | Proceed along the following steps:
| |
| | | | |
| − | <div style="padding: 5px; background: #EEEEEE;">
| + | ===(2.1) The Cenancestor's APSES Domains (1 mark)=== |
| − | #Add the secondary structure annotation to the sequence alignment in Jalview. Copy, select File → Add sequences → from Textbox and paste the sequence.
| |
| − | #Select Help → Documentation and read about Editing Alignments, Cursor Mode and Key strokes.
| |
| − | #Click on the yeast Mbp1 sequence row to select the entire row. Then use the cursor key to move that sequence directly above the 1SW6 sequence. Select the row of 1SW6 and use shift/mouse to move the sequence elements and realign them with yeast Mbp1. Refer to the alignment given in the [[Mbp1_annotation|Mbp1 annotation page]].
| |
| − | #Align the secondary structure elements with the 1SW6 sequence: Every character of 1SW6 should be matched with either E, t, H, or _. The result should be similar to the [[Mbp1_annotation|Mbp1 annotation page]]. If you need to insert gaps into all sequences in the alignment, simply drag your mouse over all row headers - movement of sequences is constrained to selected regions, the rest is locked into place to prevent inadvertent misalignments. Remember to save your project from time to time: File → save so you can reload a previous state if anything goes wrong and can't be fixed with Edit → Undo.
| |
| − | #Finally align the two CD00204 consensus sequences to their correct positions (again, refer to the [[Mbp1_annotation|Mbp1 annotation page]]).
| |
| − | #You can now consider the principles stated above and see if you can improve the alignment, for example by moving indels out of regions of secondary structure if that is possible without changing the character of the aligned columns significantly. Select blocks within which to work to leave the remaining alignment unchanged. So that this does not become tedious, you can restrict your editing to one Ankyrin repeat that is structurally defined in Swi6. You may want to open the 1SW6 structure in VMD to define the boundaries of one such repeat. You can copy and paste sections from Jalview into your assignment for documentation or export sections of the alignment to HTML (see the example below).
| |
| | </div> | | </div> |
| | | | |
| | + | Refer to your tree or the reference tree for the following two tasks. Be specific, to support your arguments, i.e. use specific branchpoints (by numbers or letters) and OTU or gene names in your arguments (see the example below). |
| | | | |
| − | <div style="padding: 5px; background: #F0F4FA; border:solid 1px #AAAAAA;"> | + | <br> |
| | + | <br> |
| | + | <div style="padding: 5px; background: #FFCC99;"> |
| | + | ;Analysis (1 mark) |
| | | | |
| − | ===(2.4.1) Editing ankyrin domain alignments - Sample===
| + | Discuss briefly how many APSES domain proteins the fungal cenancestor appears to have posessed and what evidence you see in the tre that this is so. |
| | </div> | | </div> |
| − | | + | <br> |
| − | This sample was created by
| + | |
| − | | |
| − | # Editing the alignments as described above;
| |
| − | # Copying a block of aligned sequence;
| |
| − | # Pasting it To New Alignment;
| |
| − | # Colouring the residues by Hydrophobicity and setting the colour saturation according to Conservation;
| |
| − | # Choosing File → Export Image → HTML and pasting the resulting HTML source into this Wikipage.
| |
| − | | |
| − | | |
| − | <table border="1"><tr><td>
| |
| − | <table border="0" cellpadding="0" cellspacing="0">
| |
| − | | |
| − | <tr><td colspan="6"></td>
| |
| − | <td colspan="9">10<br>|</td><td></td>
| |
| − | <td colspan="9">20<br>|</td><td></td>
| |
| − | <td colspan="9">30<br>|</td><td></td>
| |
| − | <td colspan="3"></td><td colspan="3">40<br>|</td>
| |
| − | | |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_USTMA/341-368 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#fbeef1">F</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | | |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">E</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#d3c2ee">P</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#ccaddf">T</td>
| |
| − | <td bgcolor="#ecc2d5">M</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | | |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1B_SCHCO/470-498 </td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#f7d8e0">F</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">E</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | | |
| − | <td bgcolor="#b0adfa">N</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#fcc2c4">V</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_ASHGO/465-494 </td>
| |
| − | <td>F</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#f4eef8">T</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#efc2d0">C</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | | |
| − | <td bgcolor="#e6d8f0">S</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#d3c2ee">P</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e5adc6">M</td>
| |
| − | | |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_CLALU/550-586 </td>
| |
| − | <td>G</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | | |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td>N</td>
| |
| − | <td>D</td>
| |
| − | <td>K</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | <td>S</td>
| |
| − | <td>K</td>
| |
| − | <td>F</td>
| |
| − | <td>L</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#edadbd">F</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | | |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#c6ade5">Y</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f9eef3">M</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBPA_COPCI/514-542 </td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#fbeef1">F</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fdd8da">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">E</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | | |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#ffadad">I</td>
| |
| − | <td bgcolor="#b0adfa">N</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#fcc2c4">V</td>
| |
| − | | |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_DEBHA/507-550 </td>
| |
| − | <td>I</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td>K</td>
| |
| − | <td>K</td>
| |
| − | | |
| − | <td>L</td>
| |
| − | <td>S</td>
| |
| − | <td>L</td>
| |
| − | <td>S</td>
| |
| − | <td>D</td>
| |
| − | <td>K</td>
| |
| − | <td>K</td>
| |
| − | <td>E</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | | |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | <td>A</td>
| |
| − | <td>K</td>
| |
| − | <td>F</td>
| |
| − | <td>I</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | | |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#edadbd">F</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | | |
| − | <td bgcolor="#fbadaf">V</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#c6ade5">Y</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1A_SCHCO/388-415 </td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fdd8da">V</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#fbeef1">F</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">E</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">E</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | | |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#ccaddf">T</td>
| |
| − | <td bgcolor="#ecc2d5">M</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#efc2d0">C</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | | |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_AJECA/374-403 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#f9eef3">M</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#e6d8f0">S</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | | |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">K</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_PARBR/380-409 </td>
| |
| − | <td>I</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | | |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | | |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#e6d8f0">S</td>
| |
| − | | |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | | |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">K</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | | |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_NEOFI/363-392 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#e6d8f0">S</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | | |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#fcc2c4">V</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | | |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_ASPNI/365-394 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#fbeef1">F</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | | |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#e6d8f0">S</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#fbadaf">V</td>
| |
| − | | |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#fcc2c4">V</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_UNCRE/377-406 </td>
| |
| − | <td>M</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f2d8e5">A</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | | |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">K</td>
| |
| − | | |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_PENCH/439-468 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#f9eef3">M</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#e6d8f0">S</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">Q</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#fbadaf">V</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | | |
| − | <td bgcolor="#fcc2c4">V</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBPA_TRIVE/407-436 </td>
| |
| − | | |
| − | <td>V</td>
| |
| − | <td bgcolor="#fbeef1">F</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#e6d8f0">S</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | | |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">K</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | | |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#faeef2">C</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_PHANO/400-429 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#f4eef9">W</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f4eef8">T</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | | |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | | |
| − | <td bgcolor="#c5c2fb">Q</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#ffadad">I</td>
| |
| − | <td bgcolor="#e5adc6">M</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | | |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBPA_SCLSC/294-313 </td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | | |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#ffadad">I</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">K</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | | |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBPA_PYRIS/363-392 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#f4eef9">W</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | | |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f4eef8">T</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">Q</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#ffadad">I</td>
| |
| − | <td bgcolor="#e5adc6">M</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | | |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_/361-390 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>G</td>
| |
| − | <td>V</td>
| |
| − | <td>L</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f7d8e0">F</td>
| |
| − | <td bgcolor="#f3d8e4">M</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | | |
| − | <td bgcolor="#f4eef8">T</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | | |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | | |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_ASPFL/328-364 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | | |
| − | <td>I</td>
| |
| − | <td>T</td>
| |
| − | <td>L</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f7d8e0">F</td>
| |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | <td>S</td>
| |
| − | | |
| − | <td>E</td>
| |
| − | <td>I</td>
| |
| − | <td>V</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#b0adfa">N</td>
| |
| − | <td bgcolor="#f9c2c7">L</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | | |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBPA_MAGOR/375-404 </td>
| |
| − | <td>Q</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | | |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#fbeef1">F</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#fbadaf">V</td>
| |
| − | | |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#f9c2c7">L</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#b0adfa">Q</td>
| |
| − | <td bgcolor="#c2c2ff">R</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_CHAGL/361-390 </td>
| |
| − | <td>S</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | | |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#fbadaf">V</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#f9c2c7">L</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e5adc6">M</td>
| |
| − | | |
| − | <td bgcolor="#c2c2ff">R</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_PODAN/372-401 </td>
| |
| − | <td>V</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">E</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | | |
| − | <td bgcolor="#f9c2c7">L</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#fcc2c4">V</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_LACTH/458-487 </td>
| |
| − | | |
| − | <td>F</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">Q</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | | |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#fbadaf">V</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#f9c2c7">L</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#b0adfa">Q</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | | |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_FILNE/433-460 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fdd8da">V</td>
| |
| − | | |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#fbeef1">F</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | | |
| − | <td bgcolor="#c5c2fb">E</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">E</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#ccaddf">T</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | | |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_KLULA/477-506 </td>
| |
| − | <td>F</td>
| |
| − | | |
| − | <td bgcolor="#f4eef8">T</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#fdeeee">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | | |
| − | <td bgcolor="#d3c2ee">P</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#d5c2ec">Y</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#ccaddf">T</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_SCHST/468-501 </td>
| |
| − | <td>A</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | | |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#ffd8d8">I</td>
| |
| − | | |
| − | <td>A</td>
| |
| − | <td>K</td>
| |
| − | <td>F</td>
| |
| − | <td>I</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | | |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | <td bgcolor="#edadbd">F</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#eaadc0">C</td>
| |
| − | | |
| − | <td bgcolor="#caade0">S</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_SACCE/496-525 </td>
| |
| − | <td>F</td>
| |
| − | <td bgcolor="#f4eef8">S</td>
| |
| − | | |
| − | <td bgcolor="#f2eefa">P</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#f3eef9">Y</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#eeeefe">E</td>
| |
| − | | |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | | |
| − | <td bgcolor="#f4eef8">T</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c2c2ff">K</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#ebc2d5">A</td>
| |
| − | | |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#caade0">S</td>
| |
| − | <td bgcolor="#adadff">K</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | | |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">CD00204/1-19 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c5c2fb">E</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | | |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#d8d8ff">R</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#d3c2ee">P</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#f9c2c7">L</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | | |
| − | <td bgcolor="#caade0">S</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">CD00204/99-118 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fdd8da">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | | |
| − | <td bgcolor="#eeeeff">R</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#c2c2ff">K</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | <td bgcolor="#d8d8ff">R</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#d3c2ee">P</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | | |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#f9c2c7">L</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">K</td>
| |
| − | <td bgcolor="#c5c2fb">N</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">1SW6/203-232 </td>
| |
| − | <td>L</td>
| |
| − | <td bgcolor="#eeeefe">D</td>
| |
| − | <td bgcolor="#fdeeef">L</td>
| |
| − | <td bgcolor="#eeeeff">K</td>
| |
| − | <td bgcolor="#f4eef9">W</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td bgcolor="#ffeeee">I</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f3d8e4">M</td>
| |
| − | <td bgcolor="#fbd8db">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dad8fd">N</td>
| |
| − | <td bgcolor="#f9eef3">A</td>
| |
| − | <td bgcolor="#eeeefe">Q</td>
| |
| − | <td bgcolor="#c5c2fb">D</td>
| |
| − | <td bgcolor="#d8c2e8">S</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | <td bgcolor="#cfaddc">G</td>
| |
| − | | |
| − | <td bgcolor="#dad8fd">D</td>
| |
| − | <td bgcolor="#d9c2e7">T</td>
| |
| − | <td bgcolor="#efc2d0">C</td>
| |
| − | <td bgcolor="#f7adb3">L</td>
| |
| − | <td bgcolor="#b0adfa">N</td>
| |
| − | <td bgcolor="#ffc2c2">I</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#e4adc7">A</td>
| |
| − | <td bgcolor="#adadff">R</td>
| |
| − | | |
| − | <td bgcolor="#f9c2c7">L</td>
| |
| − | <td bgcolor="#f4eef7">G</td>
| |
| − | <td bgcolor="#eeeefe">N</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">SecStruc/203-232 </td>
| |
| − | <td>t</td>
| |
| − | <td bgcolor="#f5eef6">_</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | | |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td bgcolor="#efeefd">H</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#ead8ed">_</td>
| |
| − | <td bgcolor="#ead8ed">_</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#ead8ed">_</td>
| |
| − | <td bgcolor="#f5eef6">_</td>
| |
| − | <td bgcolor="#f5eef6">_</td>
| |
| − | | |
| − | <td bgcolor="#dec2e3">_</td>
| |
| − | <td bgcolor="#d9c2e7">t</td>
| |
| − | <td bgcolor="#f5eef6">_</td>
| |
| − | <td bgcolor="#d2add8">_</td>
| |
| − | <td bgcolor="#ead8ed">_</td>
| |
| − | <td bgcolor="#dec2e3">_</td>
| |
| − | <td bgcolor="#c7c2f9">H</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | | |
| − | <td bgcolor="#c7c2f9">H</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#b3adf7">H</td>
| |
| − | <td bgcolor="#c7c2f9">H</td>
| |
| − | <td bgcolor="#f5eef6">_</td>
| |
| − | <td bgcolor="#f5eef6">_</td>
| |
| − | </tr>
| |
| − | </table>
| |
| − | </td></tr>
| |
| − | | |
| − | </table>
| |
| − | ;Aligned sequences before editing. The algorithm has placed gaps into the Swi6 helix <code>LKWIIAN</code> and the four-residue gaps before the block of well aligned sequence on the right are poorly supported.
| |
| − | | |
| − | | |
| − | <table border="1"><tr><td>
| |
| − | <table border="0" cellpadding="0" cellspacing="0">
| |
| − | | |
| − | <tr><td colspan="6"></td>
| |
| − | <td colspan="9">10<br>|</td><td></td>
| |
| − | <td colspan="9">20<br>|</td><td></td>
| |
| − | | |
| − | <td colspan="9">30<br>|</td><td></td>
| |
| − | <td colspan="3"></td><td colspan="3">40<br>|</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_USTMA/341-368 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#ffbfbf">I</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#f5d2db">F</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | | |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">E</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#c2abe8">P</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#bf99d7">T</td>
| |
| − | <td bgcolor="#e5abc5">M</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | | |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1B_SCHCO/470-498 </td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f2bfcc">F</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">E</td>
| |
| − | | |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#9d99f9">N</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | | |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#fcabae">V</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_ASHGO/465-494 </td>
| |
| − | <td>F</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | | |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#e2d2ed">T</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#ffbfbf">I</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | | |
| − | <td bgcolor="#eaabbf">C</td>
| |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#d6bfe7">S</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#c2abe8">P</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | | |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#df99b8">M</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_CLALU/550-586 </td>
| |
| − | <td>G</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td>K</td>
| |
| − | | |
| − | <td>K</td>
| |
| − | <td>E</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>L</td>
| |
| − | <td>I</td>
| |
| − | <td>S</td>
| |
| − | <td>K</td>
| |
| − | <td bgcolor="#f2bfcc">F</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | | |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#e999ad">F</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | | |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#b899df">Y</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#f0d2df">M</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBPA_COPCI/514-542 </td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#f5d2db">F</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#fcbfc1">V</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">E</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#ff9999">I</td>
| |
| − | | |
| − | <td bgcolor="#9d99f9">N</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#fcabae">V</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_DEBHA/507-550 </td>
| |
| − | <td>I</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td>K</td>
| |
| − | <td>K</td>
| |
| − | <td>L</td>
| |
| − | <td>S</td>
| |
| − | <td>L</td>
| |
| − | <td>S</td>
| |
| − | <td>D</td>
| |
| − | <td>K</td>
| |
| − | | |
| − | <td>K</td>
| |
| − | <td>E</td>
| |
| − | <td>L</td>
| |
| − | <td>I</td>
| |
| − | <td>A</td>
| |
| − | <td>K</td>
| |
| − | <td bgcolor="#f2bfcc">F</td>
| |
| − | <td bgcolor="#ffbfbf">I</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | | |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | | |
| − | <td bgcolor="#e999ad">F</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#fb999c">V</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#b899df">Y</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | | |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1A_SCHCO/388-415 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | | |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fcbfc1">V</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | | |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#f5d2db">F</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">E</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">E</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | | |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#bf99d7">T</td>
| |
| − | <td bgcolor="#e5abc5">M</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#eaabbf">C</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | | |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_AJECA/374-403 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | | |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#f0d2df">M</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | | |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#d6bfe7">S</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | | |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">K</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | | |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_PARBR/380-409 </td>
| |
| − | <td>I</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#d6bfe7">S</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | | |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">K</td>
| |
| − | | |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_NEOFI/363-392 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#d6bfe7">S</td>
| |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | | |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#fcabae">V</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | | |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_ASPNI/365-394 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#f5d2db">F</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | | |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#d6bfe7">S</td>
| |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | | |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#fb999c">V</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#fcabae">V</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | | |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_UNCRE/377-406 </td>
| |
| − | <td>M</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | | |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#eabfd3">A</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | | |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | | |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">K</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_PENCH/439-468 </td>
| |
| − | <td>T</td>
| |
| − | | |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#f0d2df">M</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#d6bfe7">S</td>
| |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | | |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">Q</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#fb999c">V</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | | |
| − | <td bgcolor="#fcabae">V</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBPA_TRIVE/407-436 </td>
| |
| − | | |
| − | <td>V</td>
| |
| − | <td bgcolor="#f5d2db">F</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#d6bfe7">S</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | | |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">K</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f4d2dc">C</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_PHANO/400-429 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#e2d2ef">W</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#e2d2ed">T</td>
| |
| − | | |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | | |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">Q</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | | |
| − | <td bgcolor="#ff9999">I</td>
| |
| − | <td bgcolor="#df99b8">M</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | | |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBPA_SCLSC/294-313 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | | |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | | |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#ff9999">I</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">K</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | | |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBPA_PYRIS/363-392 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#e2d2ef">W</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | | |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#e2d2ed">T</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | | |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">Q</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | | |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#ff9999">I</td>
| |
| − | <td bgcolor="#df99b8">M</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | | |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_/361-390 </td>
| |
| − | <td>N</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | | |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#f2bfcc">F</td>
| |
| − | <td bgcolor="#ebbfd3">M</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#e2d2ed">T</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | | |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | | |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_ASPFL/328-364 </td>
| |
| − | <td>T</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | | |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#e2d2ed">T</td>
| |
| − | <td>L</td>
| |
| − | <td>G</td>
| |
| − | <td>R</td>
| |
| − | <td>F</td>
| |
| − | | |
| − | <td>I</td>
| |
| − | <td>S</td>
| |
| − | <td>E</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td bgcolor="#ffbfbf">I</td>
| |
| − | <td bgcolor="#fcbfc1">V</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | | |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#9d99f9">N</td>
| |
| − | <td bgcolor="#f7abb2">L</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | | |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBPA_MAGOR/375-404 </td>
| |
| − | <td>Q</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | | |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#f5d2db">F</td>
| |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#fb999c">V</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#f7abb2">L</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | | |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9d99f9">Q</td>
| |
| − | <td bgcolor="#ababff">R</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_CHAGL/361-390 </td>
| |
| − | <td>S</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | | |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | | |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#fb999c">V</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#f7abb2">L</td>
| |
| − | | |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#df99b8">M</td>
| |
| − | <td bgcolor="#ababff">R</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_PODAN/372-401 </td>
| |
| − | <td>V</td>
| |
| − | | |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | | |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">E</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | | |
| − | <td bgcolor="#f7abb2">L</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#fcabae">V</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_LACTH/458-487 </td>
| |
| − | | |
| − | <td>F</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#ffbfbf">I</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">Q</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#fb999c">V</td>
| |
| − | | |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#f7abb2">L</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9d99f9">Q</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_FILNE/433-460 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fcbfc1">V</td>
| |
| − | <td bgcolor="#ffbfbf">I</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | | |
| − | <td bgcolor="#f5d2db">F</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">E</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">E</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | | |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#bf99d7">T</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | | |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_KLULA/477-506 </td>
| |
| − | <td>F</td>
| |
| − | <td bgcolor="#e2d2ed">T</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#fcd2d3">V</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#ffbfbf">I</td>
| |
| − | | |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | | |
| − | <td bgcolor="#c2abe8">P</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#c5abe5">Y</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#bf99d7">T</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | </tr>
| |
| − | | |
| − | <tr><td nowrap="nowrap">MBP1_SCHST/468-501 </td>
| |
| − | <td>A</td>
| |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | | |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>L</td>
| |
| − | <td>I</td>
| |
| − | <td>A</td>
| |
| − | <td>K</td>
| |
| − | <td bgcolor="#f2bfcc">F</td>
| |
| − | | |
| − | <td bgcolor="#ffbfbf">I</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | | |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#e999ad">F</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#e699b1">C</td>
| |
| − | <td bgcolor="#be99d9">S</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | | |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">MBP1_SACCE/496-525 </td>
| |
| − | <td>F</td>
| |
| − | <td bgcolor="#e2d2ee">S</td>
| |
| − | <td bgcolor="#ded2f2">P</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#dfd2f0">Y</td>
| |
| − | | |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#d4d2fc">E</td>
| |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#e2d2ed">T</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#ababff">K</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | | |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#e3abc6">A</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#be99d9">S</td>
| |
| − | <td bgcolor="#9999ff">K</td>
| |
| − | | |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">CD00204/1-19 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#afabfa">E</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | | |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#bfbfff">R</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#c2abe8">P</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#f7abb2">L</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | | |
| − | <td bgcolor="#be99d9">S</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">CD00204/99-118 </td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#fcbfc1">V</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d2d2ff">R</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | <td bgcolor="#ababff">K</td>
| |
| − | | |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#bfbfff">R</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#c2abe8">P</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#f7abb2">L</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | | |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">K</td>
| |
| − | <td bgcolor="#afabfa">N</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">1SW6/203-232 </td>
| |
| − | <td>L</td>
| |
| − | <td bgcolor="#d4d2fc">D</td>
| |
| − | | |
| − | <td bgcolor="#fbd2d5">L</td>
| |
| − | <td bgcolor="#d2d2ff">K</td>
| |
| − | <td bgcolor="#e2d2ef">W</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#ffd2d2">I</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#ebbfd3">M</td>
| |
| − | <td bgcolor="#f9bfc4">L</td>
| |
| − | <td bgcolor="#c2bffc">N</td>
| |
| − | <td bgcolor="#f0d2e0">A</td>
| |
| − | <td bgcolor="#d4d2fc">Q</td>
| |
| − | <td bgcolor="#afabfa">D</td>
| |
| − | | |
| − | <td bgcolor="#caabe0">S</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | <td bgcolor="#c399d4">G</td>
| |
| − | <td bgcolor="#c2bffc">D</td>
| |
| − | <td bgcolor="#cbabdf">T</td>
| |
| − | <td bgcolor="#eaabbf">C</td>
| |
| − | <td bgcolor="#f699a1">L</td>
| |
| − | <td bgcolor="#9d99f9">N</td>
| |
| − | <td bgcolor="#ffabab">I</td>
| |
| − | | |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#dd99b9">A</td>
| |
| − | <td bgcolor="#9999ff">R</td>
| |
| − | <td bgcolor="#f7abb2">L</td>
| |
| − | <td bgcolor="#e4d2ec">G</td>
| |
| − | <td bgcolor="#d4d2fc">N</td>
| |
| − | </tr>
| |
| − | <tr><td nowrap="nowrap">SecStruc/203-232 </td>
| |
| − | <td>t</td>
| |
| − | | |
| − | <td bgcolor="#e6d2e9">_</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td bgcolor="#d5d2fb">H</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | | |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td>-</td>
| |
| − | <td bgcolor="#dcbfe1">_</td>
| |
| − | <td bgcolor="#dcbfe1">_</td>
| |
| − | <td bgcolor="#dcbfe1">_</td>
| |
| − | <td bgcolor="#e6d2e9">_</td>
| |
| − | <td bgcolor="#e6d2e9">_</td>
| |
| − | | |
| − | <td bgcolor="#d2abd8">_</td>
| |
| − | <td bgcolor="#cbabdf">t</td>
| |
| − | <td bgcolor="#e6d2e9">_</td>
| |
| − | <td bgcolor="#c799cf">_</td>
| |
| − | <td bgcolor="#dcbfe1">_</td>
| |
| − | <td bgcolor="#d2abd8">_</td>
| |
| − | <td bgcolor="#b2abf7">H</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | | |
| − | <td bgcolor="#b2abf7">H</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#a199f6">H</td>
| |
| − | <td bgcolor="#b2abf7">H</td>
| |
| − | <td bgcolor="#e6d2e9">_</td>
| |
| − | <td bgcolor="#e6d2e9">_</td>
| |
| − | </tr>
| |
| − | </table>
| |
| − | </td></tr>
| |
| − | | |
| − | </table>
| |
| − | ;Aligned sequence after editing. A significant cleanup of the frayed region is possible. Now there is only one insertion event, and it is placed into the loop that connects two helices of the 1SW6 structure.
| |
| − | | |
| | | | |
| | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> |
| | | | |
| − | ===(2.5) Final analysis=== | + | ===(2.2) Unraveling your organism's APSES domains (2 marks)=== |
| | </div> | | </div> |
| | | | |
| − | | + | <br> |
| − | <div style="padding: 5px; background: #EEEEEE;"> | + | <br> |
| − | * Compare the distribution of indels in the ankyrin repeat regions of your alignments. '''Review''' whether the indels in this region are concentrated in segments that connect the helices, or if they are more or less evenly distributed along the entire region of similarity. Think about whether the assertion that ''indels should not be placed in elements of secondary structure'' has merit in your alignment. Recognize that an indel in an element of secondary structure could be interpreted in a number of different ways:
| |
| − | ** The alignment is correct, the annotation is correct too: the indel is tolerated in that particular case, for example by extending the length of an α-helix or β-strand;
| |
| − | ** The alignment algorithm has made an error, the structural annotation is correct: the indel should be moved a few residues;
| |
| − | ** The alignment is correct, the structural annotation is wrong, this is not a secondary structure element after all;
| |
| − | ** Both the algorithm and the annotation are probably wrong, but we have no data to improve the situation.
| |
| − | | |
| − | (<small>NB: remember that the structural annotations have been made for the yeast protein and might have turned out differently for the other proteins...</small>)
| |
| − | | |
| − | You should be able to analyse discrepancies between annotation and expectation in a structured and systematic way. In particular if you notice indels that have been placed into structurally annotated regions of secondary structure, you should be able to comment on whether the location of the indel has strong support from aligned sequence motifs, or whether the indel could possibly be moved into a different location without much loss in alignment quality.
| |
| − | </div> | |
| − | | |
| − | | |
| | <div style="padding: 5px; background: #FFCC99;"> | | <div style="padding: 5px; background: #FFCC99;"> |
| | ;Analysis (2 marks) | | ;Analysis (2 marks) |
| | | | |
| − | *Considering the whole alignment and your experience with editing, please note in your assignment your assessment of whether the position of indels relative to structural features of the ankyrin domains in your organism's Mbp1 protein is reliable.
| + | Assume that the phylogenetic tree for fungi is correct, and that the mixed gene tree is fundamentally correct in its overall arrangement but may have local inaccuracies due to the limited resolution of the method. You have identified the APSES domain genes of the fungal cenancestor above. Apply the expectations we have stated above to discuss briefly through what sequence of duplications and/or gene loss your organism has ended up with the APSES domains it possesses today. Make specific reference to the species tree and either your constructed tree or the [[APSES_domains_reference_tree|reference tree]]. (2 marks) |
| | + | </div> |
| | + | <br> |
| | + | |
| | | | |
| − | *CDD extends the ankyrin domain annotation beyond the 1SW6 domain boundaries. Given your assessment of conservation in that region, do you think that this is reasonable in your organisms' protein? Is there evidence for this in the alignment of the CD00204 consensus with well aligned blocks of sequence beyond the positions that match Swi6?
| + | For example the following discusion for ''Saccharomyces cerevisiae'' would be sufficient for full marks: |
| − | </div>
| + | :(Numbers refer to branchpoints of the mixed gene tree, letters to branchpoints of the species tree). There are four subclades that are shared by most current species, they branch from 129, 108, 76 and (94 + 102). For the latter case, the precise resolution appears not be well resolved, but by comparison with the species tree, we can argue that branch 102 corresponds to branch (H) and should be inserted between branchpoints 94 (corresponding to (A) ) and 96 (B) , not after branch 74. This is because the species under 95 and 102 share a common ancestor (B) that is distinct from 95. ''Saccharomyces cerevisiae'' has one gene in each of these major subclades, there is no gene loss. (Note however that there is no Dikaryomycota (2) orthologue of a Sok2 gene.) ''Saccharomyces cerevisiae'' has an additional paralogue to Sok2 that created the Phd1 gene. This is shared with ''Candida albicans''. There are three possibilites to explain this: (''i'') the gene could have been duplicated before (H) and then lost in separate, independent events after I,J,K,M and N in those species that do not possess an orthologue. (''ii'') the gene could have arisen after (N) or after (K) and then passed by horizontal gene transfer from or to ''S. cerevisiae'', or ('''iii''') the annotations of orthologues could be incorrect and some of the genes labelled SokA (Sok2 paralogues) could in fact be Phd1 orthologues; if this were the case it would require a reassessment of how much gene-loss would be necessary to explain the subclade below 108. |
| | | | |
| | + | |
| | + | |
| | | | |
| | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> | | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> |
| Line 3,730: |
Line 227: |
| | | | |
| | ;Links | | ;Links |
| − | :* [http://www.ncbi.nlm.nih.gov/blast '''BLAST'''] | + | :* [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/restricted/Baldauf_2003_PhylogenyTutorial.pdf '''Review (PDF, restricted)''' Sandra Baldauf: Phylogeny for the Faint of Heart] |
| − | :* [http://www.pir.uniprot.org/?tab=mapping '''Uniprot ID mapping''' service]
| + | :* [http://evolution.genetics.washington.edu/phylip.html '''PHYLIP''' home page] |
| − | :* [http://www.ncbi.nlm.nih.gov/sutils/blink.cgi?pid=68465419 A '''BLink''' example]
| + | :* [http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html '''PHYLIP''' Web Service at the Institut Pasteur] |
| − | :* [http://www.ebi.ac.uk/clustalw/ EBI '''CLUSTAL-W''' server] | + | :*[[Assignment_5_fallback_data|'''Fallback data''']] |
| − | :* [http://www.ebi.ac.uk/muscle/ EBI '''MUSCLE''' server] | |
| − | :* [http://www.ebi.ac.uk/t-coffee/ EBI '''T-Coffee''' server]
| |
| − | :* [http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi '''CDD'''] | |
| − | :* [http://smart.embl-heidelberg.de/ '''SMART'''] | |
| − | :* [http://www.ebi.ac.uk/thornton-srv/databases/sas/ '''SAS''']
| |
| | | | |
| − | ;Lists | + | ;APSES domain alignment |
| − | :* [[Species list]] | + | :* [[APSES_domains_MUSCLE_revised|All '''APSES domains - MUSCLE aligned''' and sequence names revised]] |
| − | :* [[Mbp1_RBM_reference_sequences|'''A page of reference sequence of Mbp1 proteins''']]
| |
| − | :* [[Mbp1_annotation|'''A page of text-based annotations for the yeast Mbp1 protein''']]
| |
| | | | |
| | + | ;Tree |
| | + | :*[[APSES_domains_reference_tree|'''APSES domains reference tree''']] |
| | | | |
| − | :'''Further reading'''
| + | |
| − | :* [http://bioinformatics.oxfordjournals.org/content/24/3/319.full Moreno-Hagelsieb & Latimer compare Reciprocal Best Match vs. a related concept: Reciprocal Smallest Distance]
| + | |
| − | | |
| − | <br> | |
| | | | |
| | <div style="padding: 5px; background: #D3D8E8; border:solid 1px #AAAAAA;"> | | <div style="padding: 5px; background: #D3D8E8; border:solid 1px #AAAAAA;"> |
| | [End of assignment] | | [End of assignment] |
| | </div> | | </div> |
| − |
| |
| − | <br>
| |
| | | | |
| | If you have any questions at all, don't hesitate to mail me at [mailto:boris.steipe@utoronto.ca boris.steipe@utoronto.ca] or post your question to the [mailto:bch441_2011@googlegroups.com Course Mailing List] | | If you have any questions at all, don't hesitate to mail me at [mailto:boris.steipe@utoronto.ca boris.steipe@utoronto.ca] or post your question to the [mailto:bch441_2011@googlegroups.com Course Mailing List] |