Difference between revisions of "Multiple Sequence Alignment"
| Line 309: | Line 309: | ||
<br> <br> | <br> <br> | ||
<div style="color: #000000; background-color:#BDC3DC; font-size:100%; text-decoration:none; border:solid 2px #999999; padding: 5px;"> | <div style="color: #000000; background-color:#BDC3DC; font-size:100%; text-decoration:none; border:solid 2px #999999; padding: 5px;"> | ||
| − | === | + | ===Editing and printing=== |
</div><br> | </div><br> | ||
<br> | <br> | ||
Revision as of 15:59, 26 October 2008
Contents
- 1 Objectives
- 2 Links
- 3 Exercises
- 4 Slides
- 4.1 Uses and problems
- 4.2 Good, poor, right, wrong - the objective functions
- 4.3 Algorithms and software tools
- 4.3.1 Slide 0016
- 4.3.2 Slide 0017
- 4.3.3 Slide 0018
- 4.3.4 Slide 0019
- 4.3.5 Slide 0020
- 4.3.6 Slide 0021
- 4.3.7 Slide 0022
- 4.3.8 Slide 0023
- 4.3.9 Slide 0024
- 4.3.10 Slide 0025
- 4.3.11 Slide 0026
- 4.3.12 Slide 0027
- 4.3.13 Slide 0028
- 4.3.14 Slide 0029
- 4.3.15 Slide 0030
- 4.3.16 Slide 0031
- 4.3.17 Slide 0032
- 4.3.18 Slide 0033
- 4.3.19 Slide 0034
- 4.3.20 Slide 0035
- 4.3.21 Slide 0036
- 4.3.22 Slide 0037
- 4.4 Editing and printing
Multiple Sequence Alignment (MSA)
Objectives
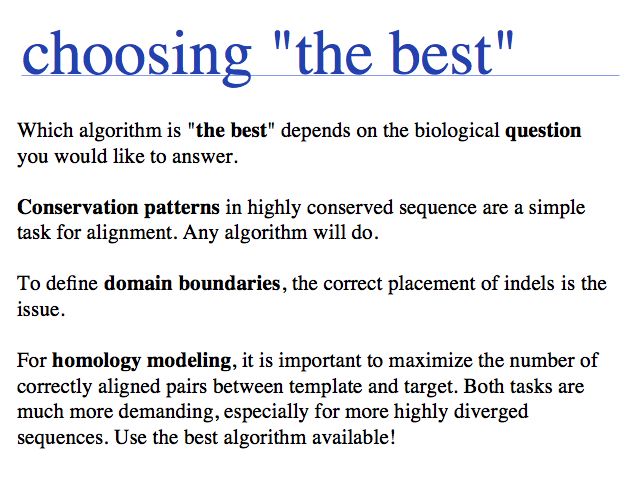
- Understand that MSA is an unsolved, difficult problem with different "best" solutions for different purposes.
- Be familiar with different biological heuristics that distinguish a "good" alignment from a "poor" alignment.
- Understand the importance of benchmarks for assessing the performance of computational tools.
- Be aware of how different biological priorities have resulted in different algorithmic strategies and know some of the available software tools that represent them.
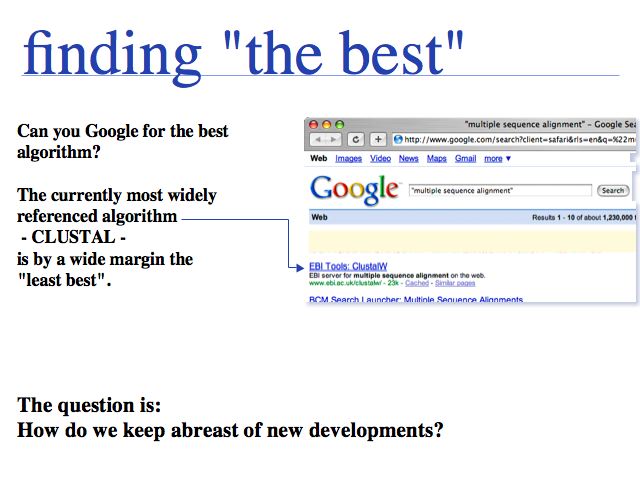
- Be aware that the most frequently used and referenced tool - CLUSTAL - is no longer state-of-the-art and know which modern tools are much better.
- Confidently be able to survey recent developments and choose an appropriate algorithm.
- Be able to perform and interpret MSAs in practice, know how to prepare input, which formats to use and what common output formats look like.
- Understand strategies to prepare input and improve alignments, based on the requirement of columnwise homology.
- Know about strategies and tools for manual editing of alignments.
Links
- Wikipedia page on Multiple Sequence Alignment
*Clustal alignments online - MUSCA
- MEME algorithm for motif discovery
- HMMER, a profile hidden Markov model tool
- Pfam domain database
- EBI TCoffee Web server
- TCoffee Web page
- EBI MUSCLE Web server
- Berkeley MUSCLE server
- Muscle homepage
- Indiana SPEM server
- Dallas PROMALS Web server
- Jalview home page
- The PFAAT homepage
- The CINEMA homepage
- Embnet BOXSHADE server
- Pasteur institute BOXSHADE server
Exercises
[...]
Slides
Uses and problems
Slide 0008
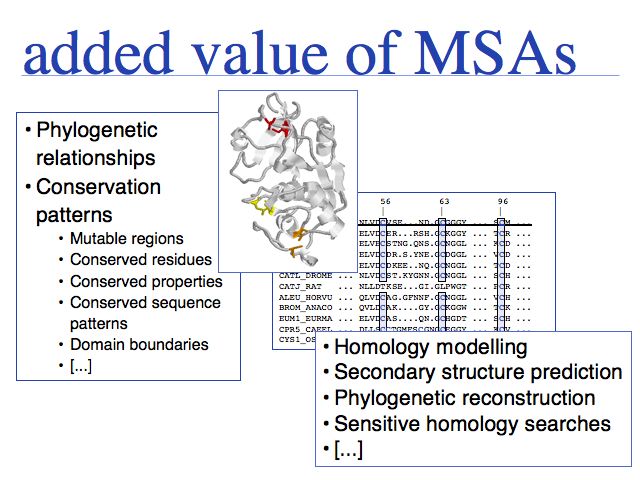
Multiple sequence alignments don't only match residues. They also give information on how strongly a residue is conserved, what it can be replaced with, which species share particular sequence patterns, and where in the sequence indels can be tolerated. An analysis of conservation even allows to distinguish between structurally and functionally conserved residues! This makes multiple sequence alignments the method of choice for many applications.
- Multiple sequence alignments are more accurate than pairwise alignments, thus they are the method of choice for starting homology modeling projects.
- Combined information from numerous sequences is invaluable for secondary structure prediction and sensitive sequence database searches.
- They contain the information needed for inferences about evolutionary relationships, i.e. the order in which particular sequence changes occurred.
Slide 0009
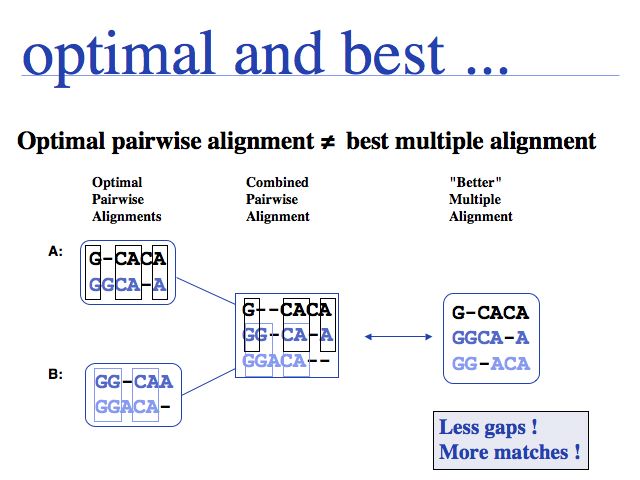
Multiple alignments cannot necessarily be constructed from pairwise alignments. Moreover, it may be impossible to merge three mutually pairwise alignments into a non-contradicting multiple alignment. However the inverse is always possible: a multiple alignment can be decomposed into pairwise alignments.
Slide 0010
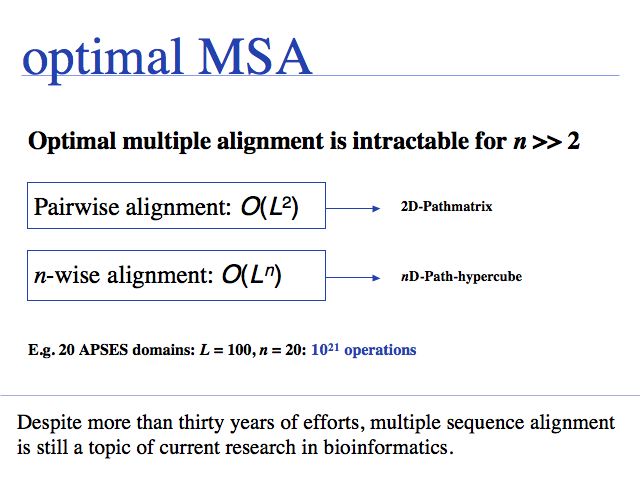
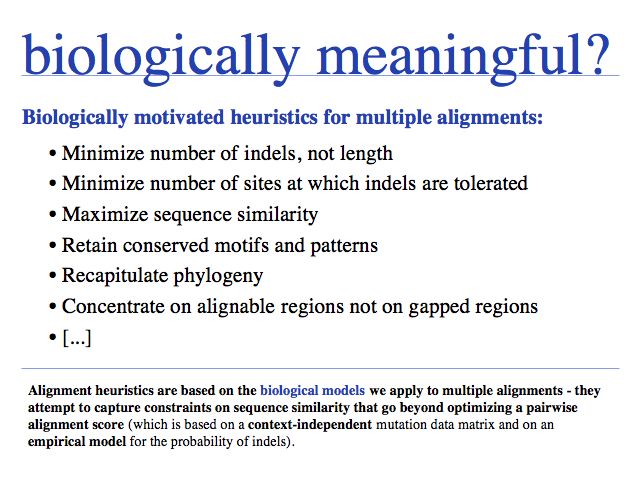
Besides being intractable, it is questionable how meaningful the objective function of optimal sequence alignments is for multiple alignments. This pair score maximizes the score derived from a mutation data matrix, for pairs of aligned residues. But - for example - the pair score does not otimize the pattern of indel placements, or whether a particular motif is well-conserved.
Good, poor, right, wrong - the objective functions
Slide 0012
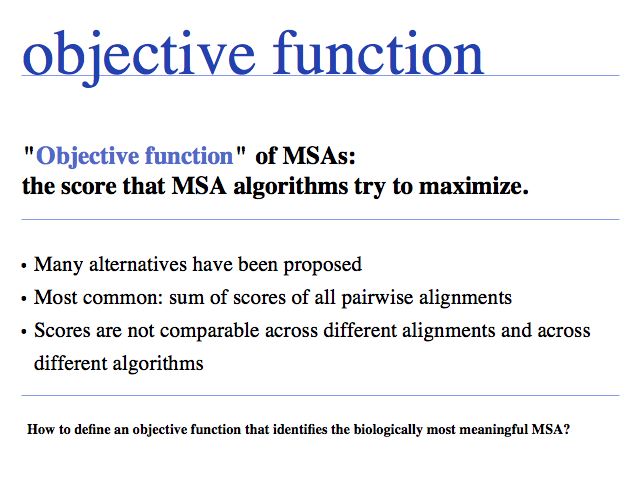
If we want an algorithm to optimize anything at all, we first must define how we can measure the quality of the result. This metric defines the target function or objective function. (Note that "objective" is not used in the sense of "unbiased" but in the sense of "target", or "goal".)
Slide 0013
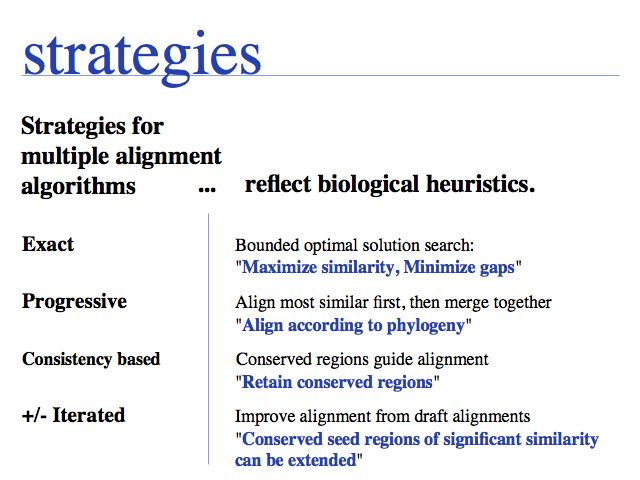
Each of the reasonable biological objectives suggests a different alignment strategy! The most modern algorithms currently available attempt to satisfy these heuristics simultaneously. Note that these are heuristics, they are not the result of some rigorously applied theory, but reflect the complex relationship between protein sequence, structure, evolution and selection.
Slide 0014
Algorithms and software tools
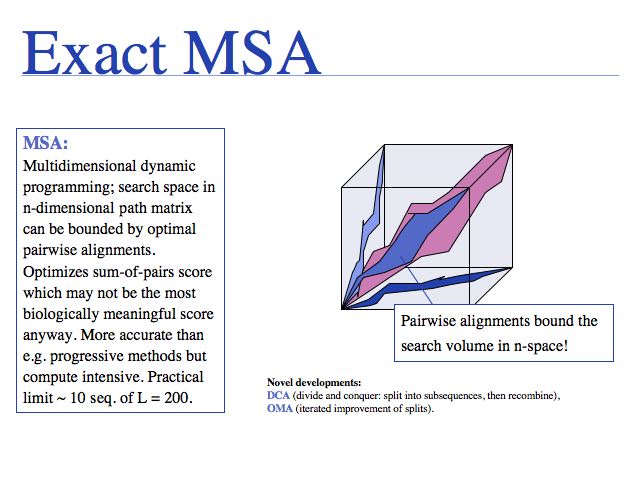
Slide 0016
Exact methods certainly have their place where it comes to analyzing and improving algorithms; they are especially of interest to computer science because high-dimensional optimal alignment is a difficult problem. However they cannot compete in terms of result-quality with modern heuristic methods.
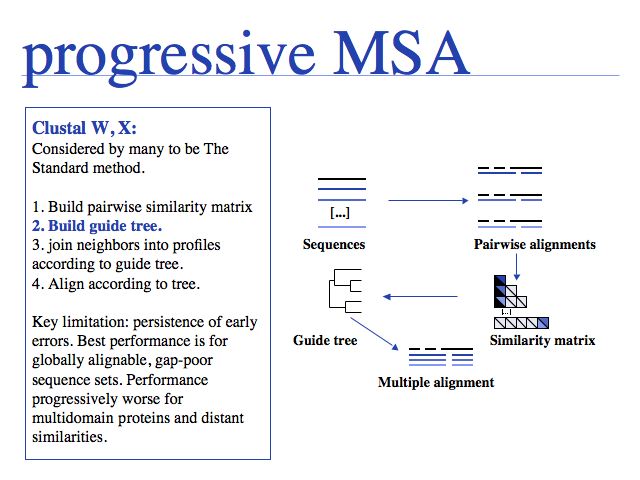
Slide 0017
Progressive alignment is one of three fundamental algorithmic approaches to MSA. The EBI offers Clustal alignments online.
Slide 0018
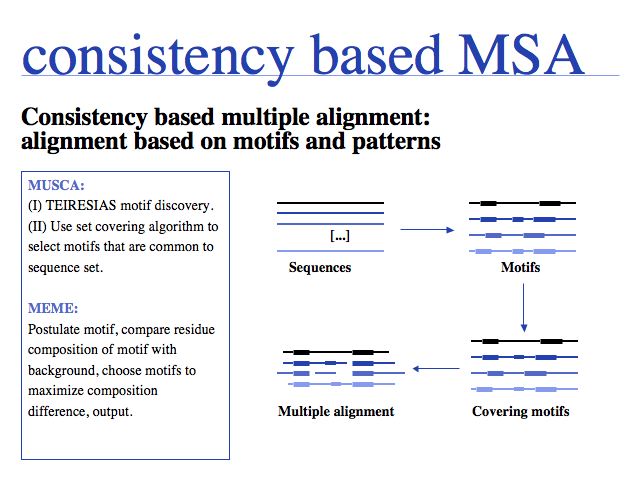
Consistency based multiple alignment is one of three fundamental algorithmic approaches to MSA. Many modern algorithms have a consistency based step included, however none of them relies solely on consistency, since problems from spurious local similarity can corrupt the alignment. MUSCA, based on the Teiresias pattern discovery algorithm is offered through IBM's Watson Labs Web server. Similarly, the MEME algorithm for motif discoverywhich is more commonly used in sequence analysis infers a motif-based alignment.
Slide 0019
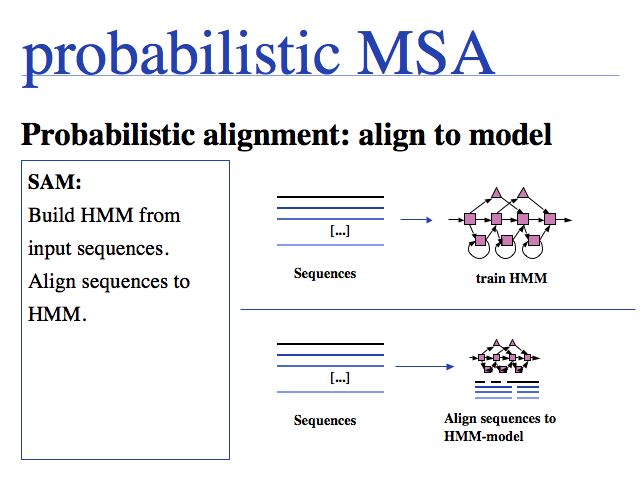
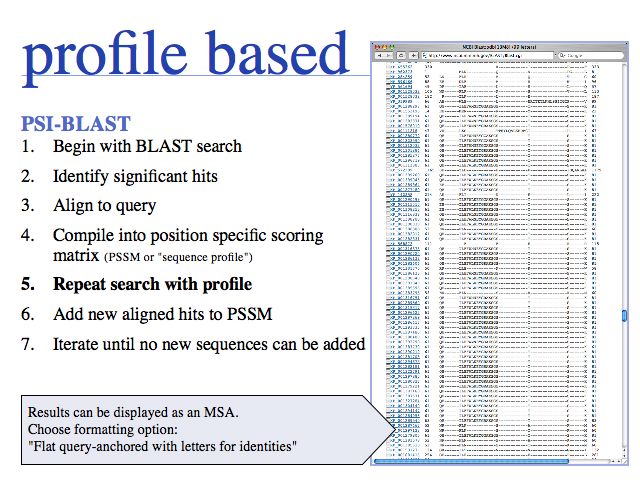
Probabilistic' multiple alignment is one of three fundamental algorithmic approaches to MSA. A statistical model of the sequences is built, then the alignment can be generated by aligning the sequences to the model. Of course, aligning sequences to a profile is a special case of this procedure: PSI BLAST can thus be used as an alignment algorithm. The most widely used algortihm is Sean Eddy's HMMER, a profile hidden Markov model tool which is also used in the generation of the Pfam domain database.
Slide 0020
Altschul et al. (1998) Nucleic Acids Research 25:3389-3402
Slide 0021
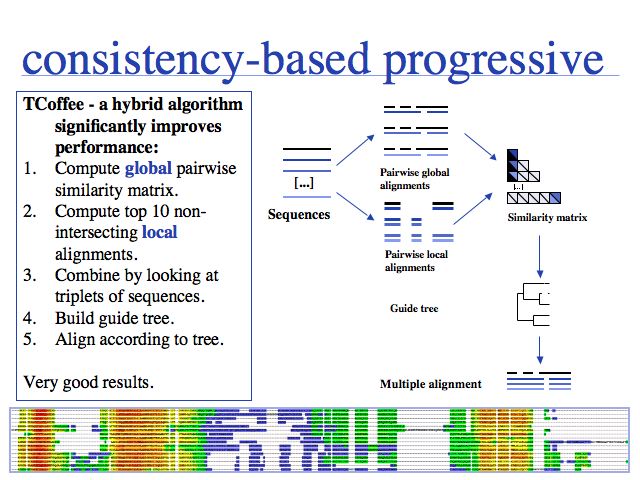
I personally rate TCoffee as the most useful and useable tool that is currently available. It is robust, fast, and gives reasonable results for many cases. Usually it is very noticeably better then CLUSTAL and I would reject any result based on CLUSTAL for that reason. Run TCoffee via the EBI TCoffee Web server which is very easy to use (although alignment size is limited;). Source code can be obtained and a local installation on UNIX machines is straightforward. The TCoffee Web page links to another Web server and also offers 3DCoffee, a variant that automatically fetches related structures and incorporates structural alignments for increased accuracy.
The inset image shows one of the useful features of TCoffee: an alignment output in which sequence is coloured according to the local quality of the alignment. This makes reliable and unreliable regions easy to spot, and immediately highlights outliers that could for example be due to sequence errors, such as frameshifts in exons. (MSA taken from the Mbp1 full-length alignment).
Slide 0022
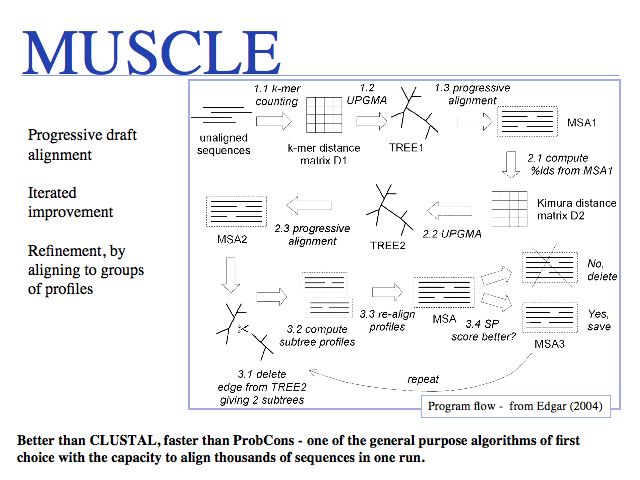
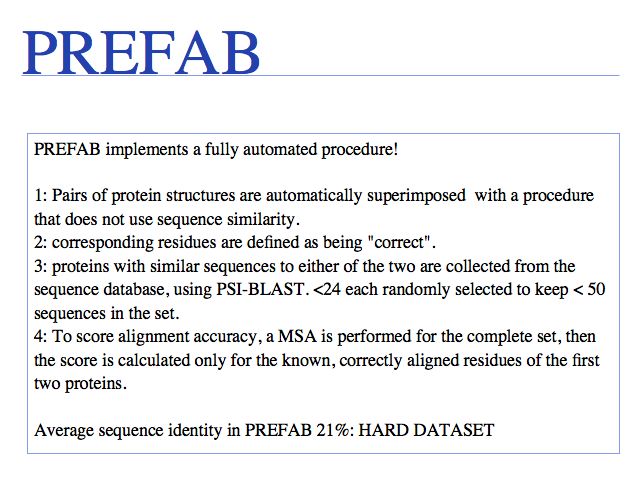
Run the MUSCLE MSAs via the EBI MUSCLE Web server which is very easy to use, or via the Berkeley MUSCLE server courtesy of Kimmen Sjolander's lab. Source code and compiled code can be obtained from the Muscle homepage and a local installation on UNIX and Windows machines is straightforward. The site also hosts the PREFAB multiple alignment benchmark.
Slide 0023
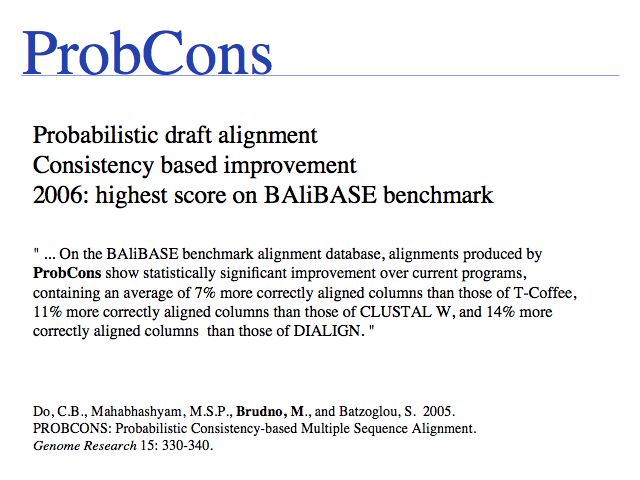
One of the best algorithms that aligns sequences without additional database information. Run it on the web via the [http://probcons.stanford.edu Stanford PROBCONS server', or download the code and install locally.
Slide 0024
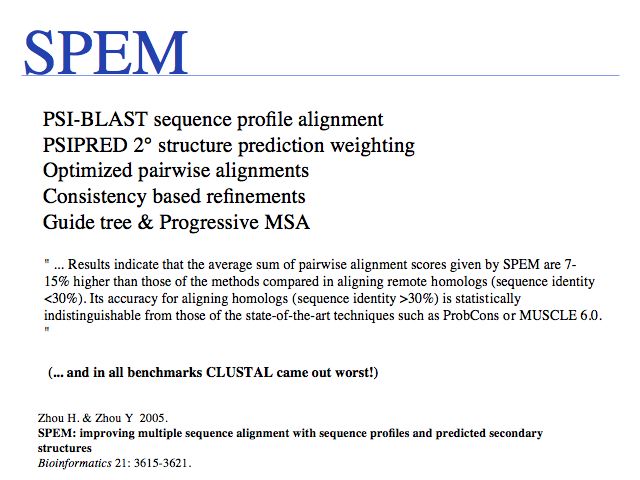
SPEM is one of the most accurate algorithms currently available, in particular for sequences of very low similarity. Run alignments via the Indiana SPEM server.
Slide 0025
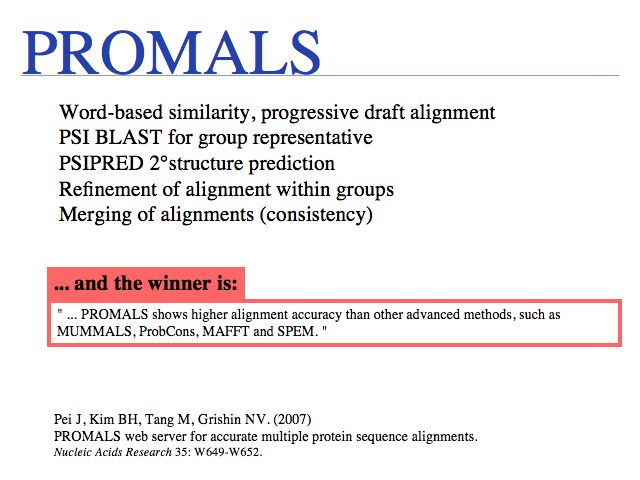
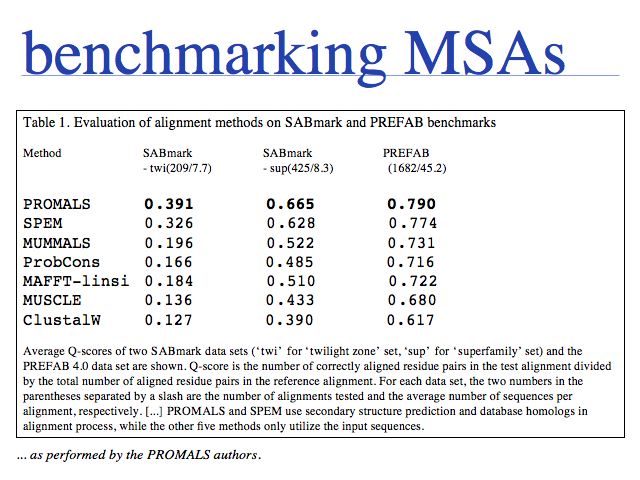
One of the latest additions to the toolkit, PROMALS is currently the most accurate MSA tool available. Run it on the Dallas PROMALS Web server. Read the PROMALS paper in the 2007 NAR Web server issue.
Slide 0026
Just what does PROMALS' improved performance mean, relative to e.g. CLUSTAL? For one, we can see a clear leap in performance through the inclusion of database information and consensus structure predictions (SPEM and PROMALS). On the other hand, regarding the SABmark superfamily dataset that is perhaps most characteristic of "typical" alignment problems with recognizeable, but low % identity, PROMALS achieves a 50% improvement relative to CLUSTAL, a 30% improvement relative to MUSCLE and ProbCons. This is much more than just statistical noise.
Slide 0027
... from the SPEM paper (Zhou & Zhou, 2005). Above ~35% pairwise sequence identity, all algortihms get it more or less right. Below ~20% pairwise sequence identity the differences are dramatic with the methods that rely on the sequences only scoring more than 20% better than CLUSTAL and SPEM outperforming CLUSTAL by about 40%.
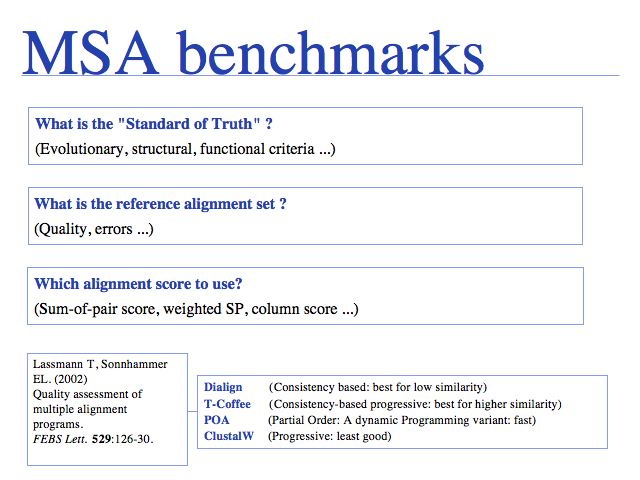
Slide 0028
How do we know that a new algorithm is better than a previous one? Benchmarks, or "Gold Standards" are an essential part of scientific hygiene. We as users must demand objective comparisons to existing methods, as referees we must require them for publication, as members of the research community we must participate in defining them and provide raw data for their construction. But we must also realize that an "arms-race" of sorts may be ensuing: as developers use the benchmarks as a training set, artificially high performance scores may be generated and performance on novel problems may degrade.
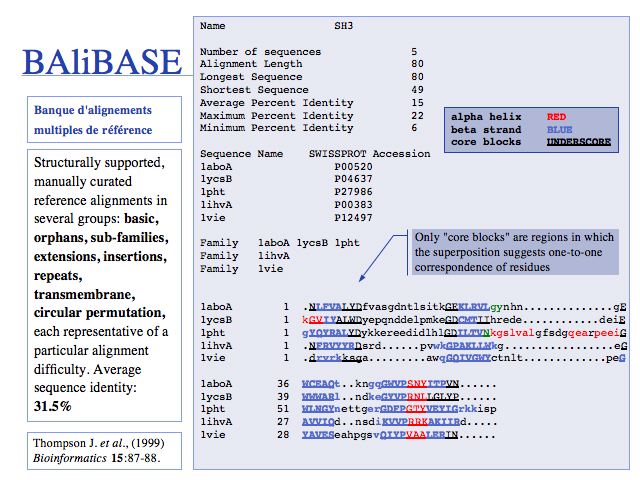
Slide 0029
Access the original BAliBASE (1999) here. Two updated versions have been created: BAliBASE 2.0 (2000) and BAliBASE 3.0. Central to BAliBASE is the concept of core blocks of alignable regions in which a pairwise correspondence of residues can be defined; outside these regions an alignment is not possible since the structural differences are too large. (BAliBASE: Thompson J. et al., (1999) Bioinformatics 15:87-88.).
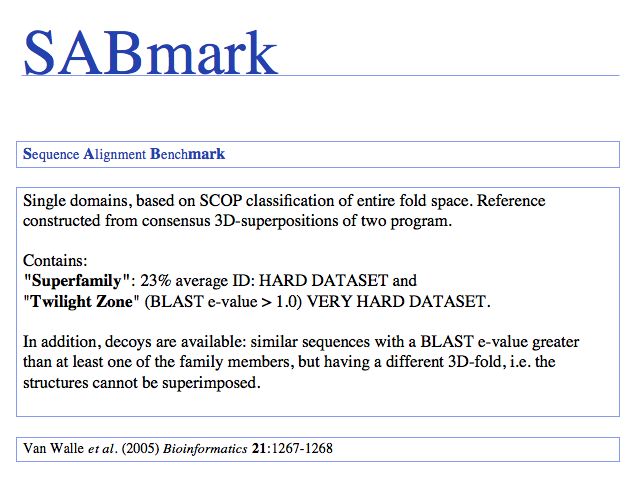
Slide 0030
SABmark: Van Walle et al. (2005) Bioinformatics 21:1267-1268. SABmark homepage.
Slide 0031
Construction of PREFAB is described in MUSCLE: Edgar (2004) Nucl Acids Res 32:1792-1797.
Slide 0032
Slide 0033
Slide 0034
Using CLUSTAL for anything but the simplest alignment problems is Cargo Cult Bioinformatics. You are doing something that may look good to the non-expert, but you can't get good results. Benchmark results have identified significant progress in the field!
"Relevance" for Google may not be the same as relevance for your work. For some applications, novelty is more important than cross-references and page-hits. For a more curated view, you can try the Wikipedia page on Multiple Sequence Alignment or the Wikiomics page. (Wikiomics is a project you should know about, but it doesn't appear to be catching on very well.)
Slide 0035
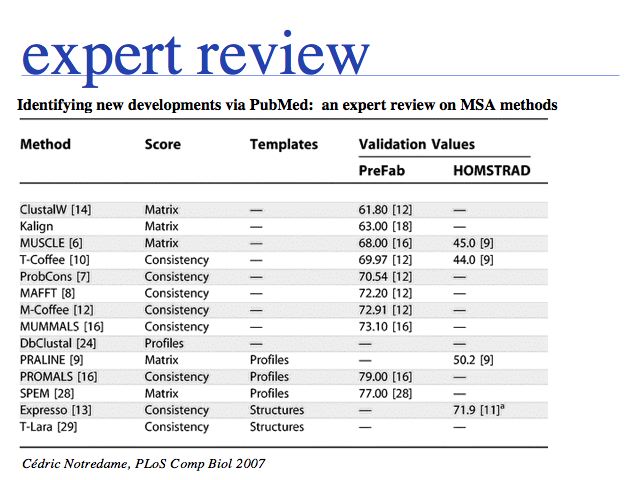
The obvious first approach is to search for a recent review. For the last year of sequence alignment literature in PubMed: search ("multiple sequence alignment"[ti] OR "multiple alignment"[ti]) AND (server OR algorithm) AND "last 1 years"[dp] or just click here. Note that not all "reviews" have been tagged by the PubMed curators as such. In the list returned in October 2008, the most recent review was found by the above search strategy. Of course, no recent review may be available, or the available reviews may not be very informative. Cedric Notredame's MSA review (2007) is technical and probably less-helpful for the non-expert, although it emphasizes the paradigm shift towards template based alignment strategies well. Edgar and Batzoglou's MSA review (2005), by the authors of MUSCLE and ProbCons, is much more readable and a good, comprehensive introduction to modern methods.
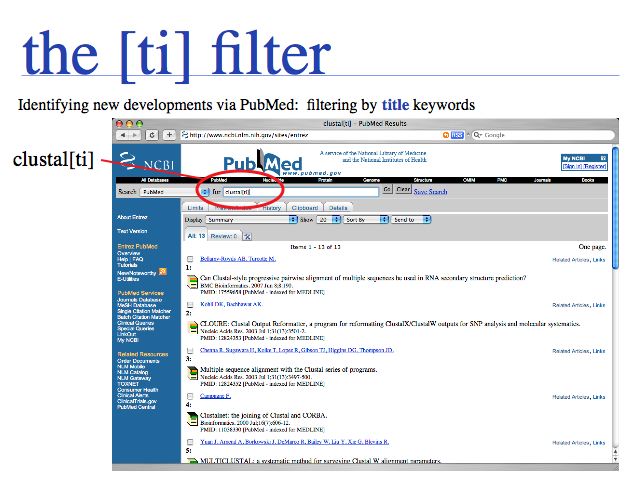
Slide 0036
An alternative and more exploratory approach is to choose a recent highly relevant article, then to use the NCBI's "Related Articles" service. This search strategy allows you to search forward in time from a particular publication. In the above example, a serch for clustal[ti] yielded a publication on CLUSTAL from 2003 ...
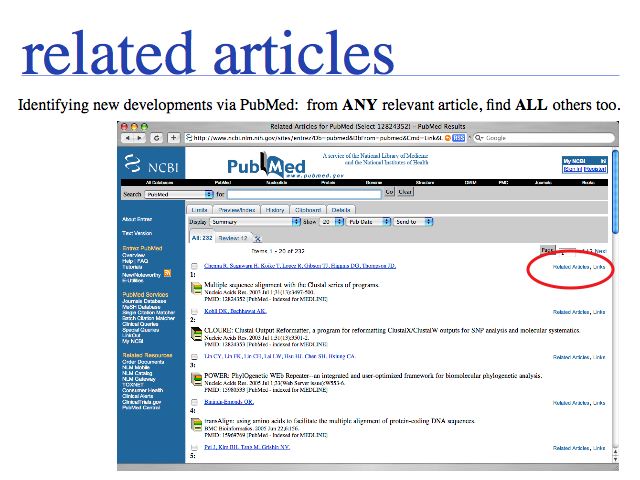
Slide 0037
... in the list of related articles (in September 2007) the article on PROMALS (2007): was number 5 in the hit-list, SPEM (2005) came as number 50.
Editing and printing
Slide 0039
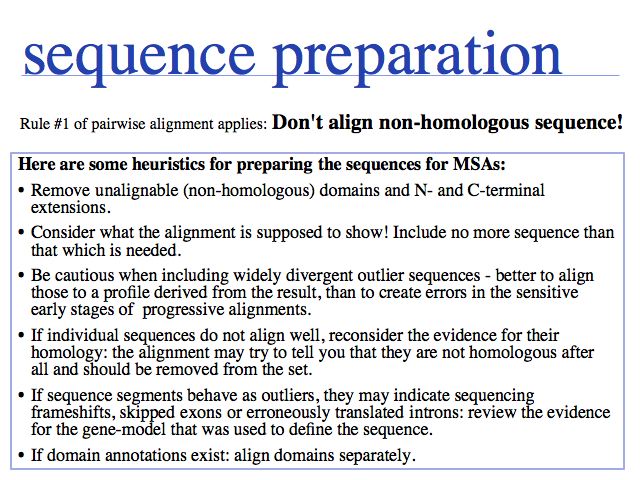
Spend some time and thought before you run the MSA to review the sequences that you are planning to align. Including un-alignable sequence will lead the algorithms astray and has the potential to degrade the entire alignment. The requirement not to align "non-hmologous" sequence should really be extended not to align (or at least: not to evaluate) sequence segments that have evolved in different context, such as in different local structural environments after insertions or deletions have occurred. The reason is: if the structural environment is not conserved, the mutation data matrix scores are irrelevant for the residues that are paired up. They may be "aligned" by the algorithm, but they are really not equivalent in structure or function, thus whether they have a good or poor similarity score is meaningless.
Slide 0040
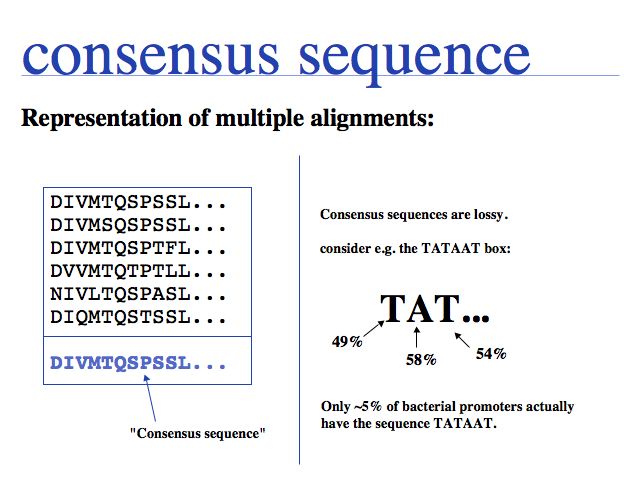
Three common formats exist for MSA results. An aligned multi FASTA file contains FASTA formatted sequences into which gap characters have been inserted. Of course, multi FASTA files can also be unaligned and they are the most common way of formatting input files for MSAs.
Slide 0041
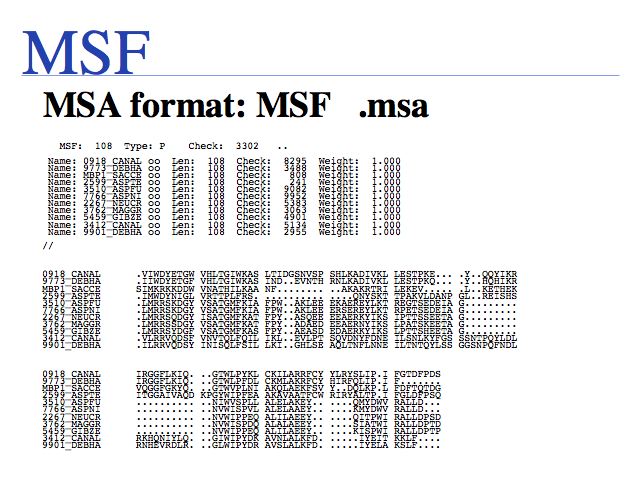
Three common formats exist for MSA results. MSF is a legacy format from the GCG package of sequence alignments, also produced by the EMBOSS tool EMMA, and supported as a valid input format for many programs. Gaps are denoted by periods and checksums are calculated for the sequences and for the alignment.
Slide 0042
Three common formats exist for MSA results. A CLUSTAL formatted alignment is the format in most common use. Take care when formatting input files to ensure the first 10 characters in your input file are unique and contain no special characters! I have seen programs break on blanks, hyphens and | (pipe). The latter is especially annoying, since the | character is used in NCBI FASTA files to separate the database identifier from the accession number. (More information at the EBI help page on formats.)
Slide 0043
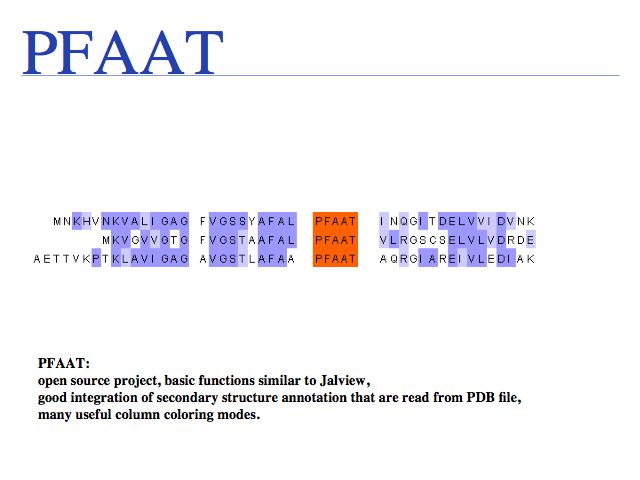
It is common and perfectly permissible to manually edit a MSA with some biologically motivated heuristic in mind as long as you document what you have done! In the early days of MSAs, editing was simply required since the results were often obviously inadequate. In all cases in which the algorithm uses only the input sequences for the alignment, this still holds true. However, regarding the more modern template-based procedures (e.g. SPEM, PROMALS or PRALINE) I would be more reluctant to edit, since we may be actively ignoring/discarding the additional information the algorithm has used.
Slide 0044
Slide 0045
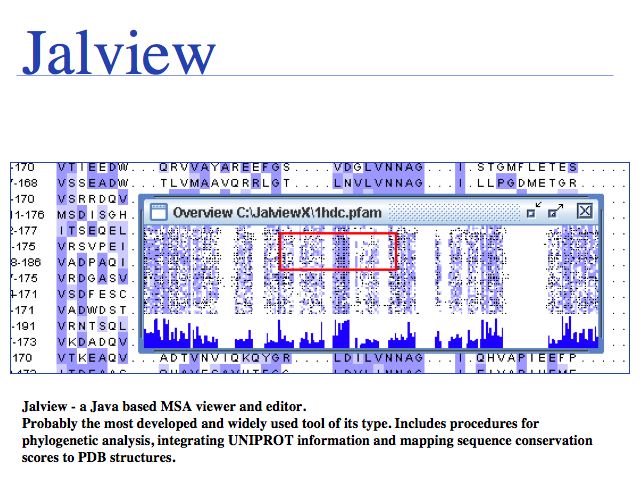
Jalview is integrated into the EBI multiple sequence alignment services, or you can access Jalview home page.
Slide 0046
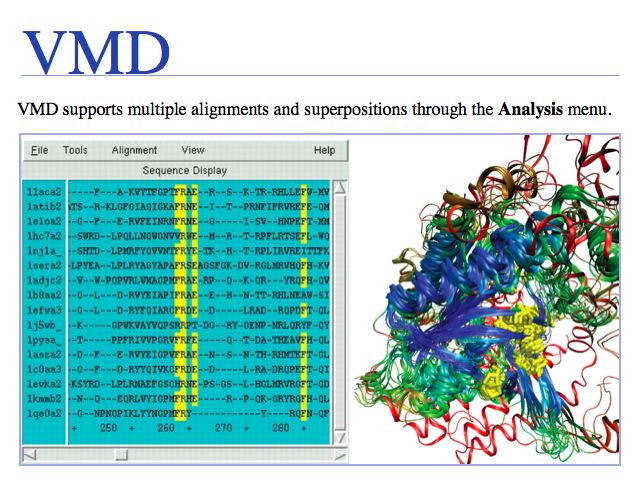
Multiple structural alignment of a representative set of the class II aminoacyl-tRNA synthetases. Structures are colored by structural conservation Qres. User-selected residues are highlighted on both the sequence and structure displays. See Eargle et al. (2006)
VMD structural alignment: Eargle et al. (2006) Bioinformatics 22:504-506
Slide 0047
Slide 0048
Slide 0049
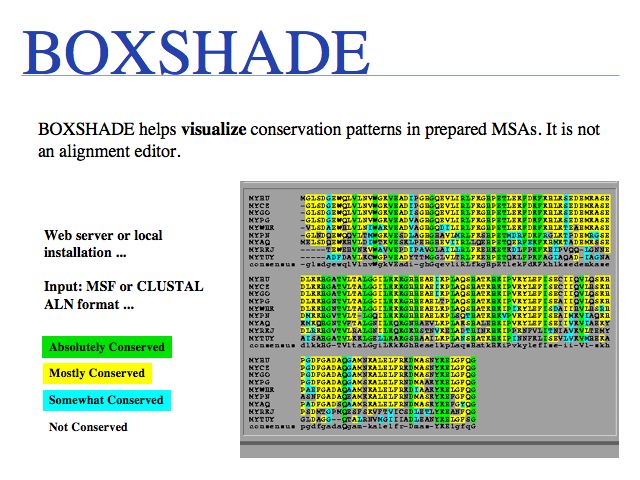
Purely for alignment visualization, run it from the Embnet BOXSHADE server or the Pasteur institute BOXSHADE server. The EMBOSS package has tools with similar functionality.
Slide 0050
Slide 0051
Slide 0052
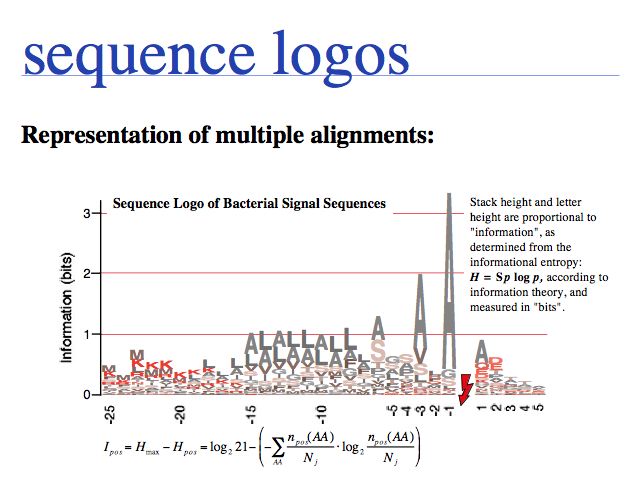
For sequence logos, see e.g. http://weblogo.berkeley.edu/ and references there.
Slide 0053