Difference between revisions of "Amino Acid Exam Questions"
Jump to navigation
Jump to search
| Line 11: | Line 11: | ||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
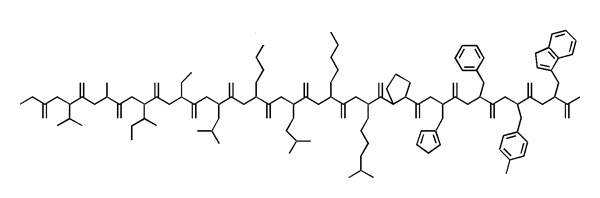
;Write the sequence of this polypeptide into your exam booklet in one-letter code. Where the sidechains are ambiguous, write all possible one letter codes for the residue in square brackets. Annotate residues that are > 80 % charged at physiological pH with a "+" or "-". | ;Write the sequence of this polypeptide into your exam booklet in one-letter code. Where the sidechains are ambiguous, write all possible one letter codes for the residue in square brackets. Annotate residues that are > 80 % charged at physiological pH with a "+" or "-". | ||
| + | </div> | ||
:''Example:'' <code>AB<sup>+</sup>CD[EFG]HIJ[KL]M<sup>-</sup>N<sup>-</sup>OPQ</code> | :''Example:'' <code>AB<sup>+</sup>CD[EFG]HIJ[KL]M<sup>-</sup>N<sup>-</sup>OPQ</code> | ||
Revision as of 16:48, 9 December 2006
- Of all data abstractions in bioinformatics, the one-letter amino acid code is the most important one. Whether we are evaluating multiple sequence alignments, database searches or mutational studies, this all requires a confident understanding of the physicochemical nature of the residues we are considering and of course knowing which letters correspond to which amino acids.
2002
- Write the sequence of this polypeptide into your exam booklet in one-letter code. Where the sidechains are ambiguous, write all possible one letter codes for the residue in square brackets. Annotate residues that are > 80 % charged at physiological pH with a "+" or "-".
- Example:
AB+CD[EFG]HIJ[KL]M-N-OPQ