Difference between revisions of "Help:ABC extensions syntax"
m (→Math markup) |
m (→Pubmedparser) |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 45: | Line 45: | ||
<nowiki>{{#pmid: 15289071 |Steipe2004}}</nowiki> | <nowiki>{{#pmid: 15289071 |Steipe2004}}</nowiki> | ||
| − | :This formats the pubmed parser output for the Cite extension; A footnote mark will be inserted ''here''{{#pmid: 15289071 |Steipe2004}} and the actual reference will appear beneath the <code><references /></code> section of the page. | + | :This formats the pubmed parser output for the Cite extension; A footnote mark will be inserted ''here''{{#pmid: 15289071 |Steipe2004}} and the actual reference will appear beneath the <code><references /></code> section of the page. Multiple citations of the same reference can use the same template, or equivalently use <code><ref name="Steipe2004" /></code>. |
| Line 78: | Line 78: | ||
|abstract= The goal of protein structure prediction is to estimate the spatial position of every atom of protein molecules from the amino acid sequence by computational methods. Depending on the availability of homologous templates in the PDB library, structure prediction approaches are categorised into template-based modelling (TBM) and free modelling (FM). While TBM is by far the only reliable method for high-resolution structure prediction, challenges in the field include constructing the correct folds without using template structures and refining the template models closer to the native state when templates are available. Nevertheless, the usefulness of various levels of protein structure predictions have been convincingly demonstrated in biological and medical applications. | |abstract= The goal of protein structure prediction is to estimate the spatial position of every atom of protein molecules from the amino acid sequence by computational methods. Depending on the availability of homologous templates in the PDB library, structure prediction approaches are categorised into template-based modelling (TBM) and free modelling (FM). While TBM is by far the only reliable method for high-resolution structure prediction, challenges in the field include constructing the correct folds without using template structures and refining the template models closer to the native state when templates are available. Nevertheless, the usefulness of various levels of protein structure predictions have been convincingly demonstrated in biological and medical applications. | ||
}} | }} | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ===Non-Pubmed DOI=== | ||
| + | |||
| + | If the article is copyrighted, don't post the PDF but use [[Template:DOI]]. Enter bibliographic details in particular <code>DOI</code> and/or <code>URL</code>. | ||
| + | |||
| + | {{DOI | ||
| + | |authors= | ||
| + | |year= | ||
| + | |title= | ||
| + | |journal= | ||
| + | |volume= | ||
| + | |pages= | ||
| + | |url= | ||
| + | |doi= | ||
| + | |abstract= | ||
| + | }} | ||
| + | |||
| + | {{DOI | ||
| + | |authors= Duncan J Murdoch, Yu-Ling Tsai & James Adcock | ||
| + | |year= 2008 | ||
| + | |title= P-Values are Random Variables | ||
| + | |journal= The American Statistician | ||
| + | |volume= 62:3 | ||
| + | |pages= 242-245 | ||
| + | |URL= | ||
| + | |doi = 10.1198/000313008X332421 | ||
| + | |file= Zhang(2012)StructurePrediction.pdf | ||
| + | |abstract= ''P''-values are taught in introductory statistics classes in a way that confuses many of the students, leading to common misconceptions about their meaning. In this article, we argue that ''p''-values should be taught through simulation, emphasizing that ''p''-values are random variables. By means of elementary examples we illustrate how to teach students valid interpretations of ''p''-values and give them a deeper understanding of hypothesis testing. | ||
| + | }} | ||
| + | |||
| + | {{Vspace}} | ||
==Cite== | ==Cite== | ||
| Line 87: | Line 120: | ||
This is the text<ref>And this is the footnote</ref>. | This is the text<ref>And this is the footnote</ref>. | ||
| + | |||
| + | Use <code><ref name="some-name">Text ...</ref></code> for the first of multiple citations of the same reference; then use <code><ref name="some-name" /ref></code> for subsequent citations. | ||
| + | |||
| + | For more options, see [https://www.mediawiki.org/wiki/Extension:Cite here]. | ||
| Line 93: | Line 130: | ||
==Misc.== | ==Misc.== | ||
| + | {{Vspace}} | ||
| + | ====raw HTML==== | ||
| + | Once <code>$wgRawHtml = true;</code> has been set, raw HTML can be used in <code><html>...</html></code> tags (https://www.mediawiki.org/wiki/Manual:$wgRawHtml). | ||
| + | |||
| + | This allows to put Google Docs in iFrames (https://wcmshelp.ucsc.edu/advanced/embedding-google/index.html): | ||
| + | |||
| + | <html> | ||
| + | <iframe src="https://docs.google.com/presentation/d/e/2PACX-1vTSqtVMXf2OC1k787ALJ43qpMwyaRy6pGBqCRZGjUMW7SapDyhLfdYZ9OpXk1quvcLthyVnvWlcrV6S/embed?start=false&loop=false&delayms=9999999" frameborder="0" width="360" height="509" allowfullscreen="true" mozallowfullscreen="true" webkitallowfullscreen="true"></iframe> | ||
| + | <img src="https://docs.google.com/drawings/d/e/2PACX-1vRYfclMZ7ovR26i2KAN8bbzmBWApImKH4tPuk66ZYwkU1H9-zy0BkuS_JFrPjwpcBqwCriY2YDCwaWH/pub?w=480&h=360"> | ||
| + | </html> | ||
| + | |||
| + | ====CC-BY==== | ||
| + | <code><nowiki>{{CC-BY}}</nowiki></code> | ||
| + | {{CC-BY}} | ||
| + | |||
| + | |||
| + | ====PDFInclude==== | ||
| + | <pdf>http://steipe.biochemistry.utoronto.ca/abc/CourseMaterials/BCH441/02-SequenceData_2014.pdf</pdf> | ||
| + | <pdf width="300px" height="400px">http://steipe.biochemistry.utoronto.ca/abc/CourseMaterials/BCH441/02-SequenceData_2014.pdf</pdf> | ||
| + | |||
====Math markup==== | ====Math markup==== | ||
| Line 98: | Line 155: | ||
| − | <math>H = - \sum_{i=0}^n p_i \log_{ | + | <math>H = - \sum_{i=0}^n p_i \log_{2} p_i</math><br /> |
:see: http://meta.wikimedia.org/wiki/Help:Formula | :see: http://meta.wikimedia.org/wiki/Help:Formula | ||
| + | |||
====Inserting a Web-site box==== | ====Inserting a Web-site box==== | ||
| Line 106: | Line 164: | ||
{{WWW|WWW_UniProt}} | {{WWW|WWW_UniProt}} | ||
| + | |||
| + | ====Inserting an emphasis box==== | ||
| + | <nowiki><div class="emphasis-box">Getting the data model right is the key to eternal bliss.</div></nowiki> | ||
| + | <div class="emphasis-box">Getting the data model right is the key to eternal bliss.</div> | ||
| + | |||
| + | |||
| + | | ||
====Inserting a Web-link box==== | ====Inserting a Web-link box==== | ||
| Line 111: | Line 176: | ||
<div class="reference-box">[http://www.ncbi.nlm.nih.gov '''The NCBI''']</div> | <div class="reference-box">[http://www.ncbi.nlm.nih.gov '''The NCBI''']</div> | ||
| + | ====Linking text to '''R''' documentation ==== | ||
| + | <table style="border:solid 1px #000000"> | ||
| + | <tr> | ||
| + | <th> </th> | ||
| + | <th> </th> | ||
| + | <th style="text-align:left;"><code>Code</code></th> | ||
| + | <th> </th> | ||
| + | <th style="text-align:left;">Result</th> | ||
| + | </tr><tr> | ||
| + | <tr> | ||
| + | <td style="text-align:right;">base function only:</td> | ||
| + | <td> </td> | ||
| + | <td><code><nowiki>{{R|mean()}}</nowiki></code></td> | ||
| + | <td> </td> | ||
| + | <td>{{R|mean()}}</td> | ||
| + | </tr><tr> | ||
| + | <td style="text-align:right;">base function with different linked text:</td> | ||
| + | <td> </td> | ||
| + | <td><code><nowiki>{{R|mean()|the '''mean()''' function}}</nowiki></code></td> | ||
| + | <td> </td> | ||
| + | <td>{{R|mean()|the '''mean()''' function}}</td> | ||
| + | </tr><tr> | ||
| + | <td style="text-align:right;">package (note: no parentheses ):</td> | ||
| + | <td> </td> | ||
| + | <td><code><nowiki>{{R|knitr}}</nowiki></code></td> | ||
| + | <td> </td> | ||
| + | <td>{{R|knitr}}</td> | ||
| + | </tr><tr> | ||
| + | <td style="text-align:right;">function in non-base package:</td> | ||
| + | <td> </td> | ||
| + | <td><code><nowiki>{{R|graphics|par()}}</nowiki></code></td> | ||
| + | <td> </td> | ||
| + | <td>{{R|graphics|par()}}</td> | ||
| + | </tr><tr> | ||
| + | <td style="text-align:right;">function with different text:</td> | ||
| + | <td> </td> | ||
| + | <td><code><nowiki>{{R|grDevices|colorRamp()|colorRampPalette()}}</nowiki></code></td> | ||
| + | <td> </td> | ||
| + | <td>{{R|grDevices|colorRamp()|colorRampPalette()}}</td> | ||
| + | </tr> | ||
| + | </table> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | Create link to online '''R''' documentation. If a name is followed by <code>()</code> it's a function, otherwise a package. If a package is given alone, link to the package documentation, otherwise to the package/function page. The ''base'' package is default, other packages can be chosen with the template's second parameter. If a third parameter is present, that will be the linked text. All documentation is pulled from [http://www.rdocumentation.org/ '''rdocumentation.org'''] and this should include CRAN as well as bioconductor packages. | ||
====Linking text to Wikipedia==== | ====Linking text to Wikipedia==== | ||
Latest revision as of 16:33, 25 February 2019
Extension Syntax
This is a synopsis of special extensions and template syntax on this Wiki.
LST
Using this markup, you can arbitrarily label sections from other pages and transclude them into the current page. This is not just convenient, it is essential to avoid duplicating text that will not get consistently updated if it exists multiple times on the Wiki.
- LST (Labeling a section for transclusion)
<section begin=section-name /> ... <section end=section-name />
- LST (Transcluding a labeled section)
{{#lst:Page-name|section-name}}
- Example
Entia non sunt ponenda praeter necessitatem. (Occam)
Entia non sunt ponenda praeter necessitatem. (Occam)
Syntax Highlight
<source lang="R">y <- as.character("yadda"); 2* y #yadda yadda</source>
y <- as.character("yadda"); 2* y #yadda yadda- lang="??"
- line="GESHI_NORMAL_LINE_NUMBERS|GESHI_FANCY_LINE_NUMBERS"
- line start="??"
- highlight="??"
- enclose="pre|div|none" ... (div induces wrapping))
Pubmedparser
{{#pmid:15289071}}
- This inserts the article information in a
<div>, formatted by Template:Pubmed.
| Steipe (2004) Consensus-based engineering of protein stability: from intrabodies to thermostable enzymes. Meth Enzymol 388:176-86. (pmid: 15289071) |
{{#pmid: 15289071 |Steipe2004}}
- This formats the pubmed parser output for the Cite extension; A footnote mark will be inserted here[1] and the actual reference will appear beneath the
<references />section of the page. Multiple citations of the same reference can use the same template, or equivalently use<ref name="Steipe2004" />.
Non-Pubmed pdf
If the article is not on PubMed use Template:PDF. Upload the article to the repository, enter bibliographic details and make sure file the filename is unique with full extension.
{{PDF
|authors=
|year=
|title=
|journal=
|volume=
|pages=
|URL=
|doi=
|file=
|abstract=
}}
| Ambrish, R. & Zhang, Y. (2012) Protein Structure Prediction. Encyclopedia of Life Sciences |
| (pmid: None) [ Source URL ] Abstract |
Non-Pubmed DOI
If the article is copyrighted, don't post the PDF but use Template:DOI. Enter bibliographic details in particular DOI and/or URL.
{{DOI
|authors=
|year=
|title=
|journal=
|volume=
|pages=
|url=
|doi=
|abstract=
}}
| Duncan J Murdoch, Yu-Ling Tsai & James Adcock (2008) P-Values are Random Variables. The American Statistician 62:3:242-245. (pmid: None) |
| [ DOI ] Abstract |
Cite
- Inserting a footnote
- A note in :<ref>:</ref> tags will insert a footnote mark, linked to the text in the <references /> section of the page.
This is the text<ref>And this is the footnote:</ref>.
This is the text[2].
Use <ref name="some-name">Text ...</ref> for the first of multiple citations of the same reference; then use <ref name="some-name" /ref> for subsequent citations.
For more options, see here.
Misc.
raw HTML
Once $wgRawHtml = true; has been set, raw HTML can be used in <html>...</html> tags (https://www.mediawiki.org/wiki/Manual:$wgRawHtml).
This allows to put Google Docs in iFrames (https://wcmshelp.ucsc.edu/advanced/embedding-google/index.html):
CC-BY
{{CC-BY}}
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
PDFInclude
<pdf>http://steipe.biochemistry.utoronto.ca/abc/CourseMaterials/BCH441/02-SequenceData_2014.pdf</pdf> <pdf width="300px" height="400px">http://steipe.biochemistry.utoronto.ca/abc/CourseMaterials/BCH441/02-SequenceData_2014.pdf</pdf>
Math markup
<math>H = - \sum_{i=0}^n p_i \log_{2} p_i</math>
Failed to parse (MathML with SVG or PNG fallback (recommended for modern browsers and accessibility tools): Invalid response ("Math extension cannot connect to Restbase.") from server "https://wikimedia.org/api/rest_v1/":): {\displaystyle H = - \sum_{i=0}^n p_i \log_{2} p_i}
Inserting a Web-site box
Boxes for Web site summaries, image and abstract
{{WWW|WWW_UniProt}}
| UniProt [ link ] [ page ] Expand... | 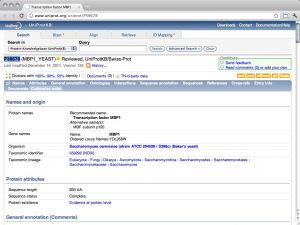 |
Inserting an emphasis box
<div class="emphasis-box">Getting the data model right is the key to eternal bliss.</div>
Inserting a Web-link box
<div class="reference-box">[http://www.ncbi.nlm.nih.gov '''The NCBI''']</div>
Linking text to R documentation
Code |
Result | |||
|---|---|---|---|---|
| base function only: | {{R|mean()}} |
mean() |
||
| base function with different linked text: | {{R|mean()|the '''mean()''' function}} |
the mean() function |
||
| package (note: no parentheses ): | {{R|knitr}} |
knitr |
||
| function in non-base package: | {{R|graphics|par()}} |
par() |
||
| function with different text: | {{R|grDevices|colorRamp()|colorRampPalette()}} |
colorRampPalette() |
Create link to online R documentation. If a name is followed by () it's a function, otherwise a package. If a package is given alone, link to the package documentation, otherwise to the package/function page. The base package is default, other packages can be chosen with the template's second parameter. If a third parameter is present, that will be the linked text. All documentation is pulled from rdocumentation.org and this should include CRAN as well as bioconductor packages.
Linking text to Wikipedia
{{WP|Mutual information|'''Mutual Information'''}}
Collapsible elements
See: Manual: Collapsible elements
<div class="mw-collapsible mw-collapsed" data-expandtext="Expand" data-collapsetext="Collapse" style="width:50%; "> * Visible text <div class="mw-collapsible-content" style="padding:10px;"> * Collapsed text 1 * Collapsed text 2 * Collapsed text 3 </div> </div>